論文

Shao X., Hsiao S.W., Xiao C., Zhou H., Tanaka Y., Macpherson T., Shichino Y., Iwasaki S., Hasegawa E., and Doi M.*:
“Translational regulation of GAD1 identified by circadian and light-responsive ribosome-bound transcriptome analysis in the mouse hypothalamic suprachiasmatic nucleus”
BioRxiv. (2026)
“Translational regulation of GAD1 identified by circadian and light-responsive ribosome-bound transcriptome analysis in the mouse hypothalamic suprachiasmatic nucleus”
BioRxiv. (2026)

Kawamoto N. and Iwasaki S.*:
“Translating the green code: Ribo-Seq for photosynthetic eukaryotes in focus”
J Biochem. (2026)
“Translating the green code: Ribo-Seq for photosynthetic eukaryotes in focus”
J Biochem. (2026)

Koch A.#, Tomuro K.#, Wakigawa T.#, Morisaki T., Iwasaki S.*, and Stasevich TJ.*:
“Bridging single-molecule and genome-wide studies of cellular mRNA translation”
RNA. (2026)
“Bridging single-molecule and genome-wide studies of cellular mRNA translation”
RNA. (2026)

Zhang R.#, Mayer L.#, Hikida H., Shichino Y., Mito M., Willemsen A., Iwasaki S.*, Ogata H.*:
“A giant virus forms a specialized subcellular environment within its amoeba host for efficient translation”
Nat Microbiol. 11(2):584-596 (2026)
BioRxiv. (2024)
Press Release (RIKEN)
Press Release (Kyoto University)
“A giant virus forms a specialized subcellular environment within its amoeba host for efficient translation”
Nat Microbiol. 11(2):584-596 (2026)
BioRxiv. (2024)
Press Release (RIKEN)
Press Release (Kyoto University)

Hosogane M*., Sue-yi S., Hatano A., Huang X., Shichino Y., Watanabe Y., Iwasaki S., Matsumoto M., and Nakayama K*.:
“Variant-specific interaction of kinectin 1 with the multi–tRNA synthetase complex regulates ER sheet organization”
iScience. 29(1):114303 (2026)
“Variant-specific interaction of kinectin 1 with the multi–tRNA synthetase complex regulates ER sheet organization”
iScience. 29(1):114303 (2026)

Iwasaki W*., Kashiwagi K., Sakamoto A., Nishimoto M., Takahashi M., Machida K., Imataka H., Matsumoto A., Shichino Y., Iwasaki S., Imami K., and Ito T*.:
“Structural insights into the role of eIF3 in translation mediated by the HCV IRES”
Proc Natl Acad Sci USA. 122(49):e2505538122 (2025)
Press Release (RIKEN)
“Structural insights into the role of eIF3 in translation mediated by the HCV IRES”
Proc Natl Acad Sci USA. 122(49):e2505538122 (2025)
Press Release (RIKEN)

Ichii M.#, Ishigami Y.#, Yao X., Li J., Sato J., Okada Y., Kimura A.P., Mito M., Iwasaki S., Kurihara Y., Noda T., Araki K., Ohtsuka M., Ohazama S., Maita H., Suzuki T., and Nakagawa S*.:
“Revisiting the in vivo functions of coilin in mice”
BioRxiv. (2025)
“Revisiting the in vivo functions of coilin in mice”
BioRxiv. (2025)

Han P., Mito M., Yamashita A., Ito T., and Iwasaki S.*:
“Sense codon-misassociated eRF1 elicits ribosome stalling and induction of quality control”
BioRxiv. (2025)
“Sense codon-misassociated eRF1 elicits ribosome stalling and induction of quality control”
BioRxiv. (2025)

Kaida D.*, Inagaki Y., Yaku K., Sasaki K., Kuwata-Ikeda S., Koike S., Shima M., Iwasaki S., Inobe T., Nakagawa T., Yoshida M., Ishigami K., and Katsuta R.:
“CGK733 binds to adenine nucleotide translocator 2 and modulates mitochondrial function and protein translation”
BioRxiv. (2025)
“CGK733 binds to adenine nucleotide translocator 2 and modulates mitochondrial function and protein translation”
BioRxiv. (2025)

Wakigawa T.#, Mito M.#, Ando Y.#, Yamashiro H., Tomuro K., Tani H., Tomizawa K., Chujo T., Nagao A., Suzuki Ta., Nureki O., Wei FY., Shichino Y., Itoh Y., Suzuki Ts., and Iwasaki S.*:
“Monitoring the complexity and dynamics of mitochondrial translation”
Mol Cell. 85(22):4279-4297.e8 (2025)
BioRxiv. (2024)
Press Release (RIKEN)
Press Release (Tohoku University)
Press Release (Kumamoto University)
Press Release (University of Tsukuba)
Press Release (The University of Tokyo)
Press Release (The University of Tokyo)
“Monitoring the complexity and dynamics of mitochondrial translation”
Mol Cell. 85(22):4279-4297.e8 (2025)
BioRxiv. (2024)
Press Release (RIKEN)
Press Release (Tohoku University)
Press Release (Kumamoto University)
Press Release (University of Tsukuba)
Press Release (The University of Tokyo)
Press Release (The University of Tokyo)

Li ST., Kamada K., Honda A., Seino J., Matsuda T., Suzuki Tak., Dohmae N., Shichino Y., Iwasaki S., Noda Y., Costanzo M., Boone C., and Suzuki Tad.*:
“LLP1 is a pyrophosphatase involved in homeostasis/quality control of dolichol-linked oligosaccharide”
J Cell Biol. 224(11):e202501239 (2025)
Press Release (RIKEN)
“LLP1 is a pyrophosphatase involved in homeostasis/quality control of dolichol-linked oligosaccharide”
J Cell Biol. 224(11):e202501239 (2025)
Press Release (RIKEN)
Świrski M.I.*, Tierney J.A.S.*, Albà M.M., Andreev D.E., Aspden J.L., Atkins J.F., Bassani-Sternberg M., Berry M.J., Biffo S., Boris-Lawrie K., Borodovsky M., Brierley I., Brook M., Brunet M.A., Bujnicki J.M., Caliskan N., Calviello L., Carvunis A.R., Cate J.H.D., Cenik C., Chang K.Y., Chen Y., Chothani S., Choudhary J.S., Clark P.L., Clauwaert J., Cooley L., Dassi E., Dean K., Diaz J.J., Dieterich C., Dikstein R., Dinman J.D., Dmitriev S.E., Dontsova O.A., Dunham C.M., Eswarappa S.M., Farabaugh P.J., Faridi P., Fierro-Monti I., Firth A.E., Gatfield D., Gebauer F., Gelfand M.S., Gray N.K., Green R., Hill C.H., Hou Y.M., Hübner N., Ignatova Z., Ivanov P.,Iwasaki S., Johnson R., Jomaa A., Jovanovic M., Jungreis I., Kellis M., Kieft J.S., Kochetov A.V., Koonin E.V., Korostelev A.A., Kufel J., Kulakovskiy I.V., Kurian L., Lafontaine D.L.J., Larsson O., Loughran G., Lukeš J., Mariotti M., Martens-Uzunova E.S., Martinez T.F., Matsumoto A., McManus J., Medenbach J., Melnikov S.V., Menschaert G., Merchante C., Mikl M., Miller W.A., Mühlemann O., Namy O., Nedialkova D.D., Nosek J., Orchard S., Ozretić P., Pertea M., Pervouchine D.D., Romão L., Ron D., Roucou X., Rubtsova M.P., Ruiz-Orera J., Saghatelian A., Salzberg S.L., Seale L.A., Seoighe C., Sergiev S.V., Shah P., Shirokikh N., Slavoff S.A., Sonenberg N., Stasevich T.J., Szczesny R.J., Tamm T., Tchórzewski M., Topisirovic I., Tremblay M.L., Tuller T., Ulitsky I., Valášek L.S., Damme P.V., Viero G., Vizcaino J.A., Vogel C., Wallace E.W.J., Weissman J.S., Westhof E., Whiffin E., Wilson D.N., Xie Z., Yewdell J.W., Yordanova M.M., Yu C.H., Yurchenko V., Zagrovic B., TRANSLACORE, Valen E.*, and Baranov P.V.*:
“Translon: a single term for translated regions”
Nat Methods. 22(10):2002-2006 (2025)
“Translon: a single term for translated regions”
Nat Methods. 22(10):2002-2006 (2025)

Shichino Y.*#, Mito M.#, Kinugasa Y., Wakigawa T., Yamashita A., Mishima Y., Imai Y., and Iwasaki S.*:
“Ultra-parallel ribosome profiling platform with RNA-dependent RNA amplification”
BioRxiv. (2025)
“Ultra-parallel ribosome profiling platform with RNA-dependent RNA amplification”
BioRxiv. (2025)

Wakigawa T.#, Fujita T.#, Kawamoto N.#, Kurihara Y., Hirose Y., Hirayama T., Toh H., Kuriyama T., Hashimoto A., Matsuura-Suzuki E., Mochida K., Yoshida M., Matsui M., and Iwasaki S.*:
“Synchronization between chloroplastic and cytosolic protein synthesis for photosynthesis complex assembly”
BioRxiv. (2025)
“Synchronization between chloroplastic and cytosolic protein synthesis for photosynthesis complex assembly”
BioRxiv. (2025)

Tomuro K., Akaiwa T., Harada A., Kobayashi K., Shiina N., Kawaguchi K.*, Shichino Y.*, and Iwasaki S.*:
“Sequence grammar and dynamics of subcellular translation revealed by APEX-Ribo-Seq.”
BioRxiv. (2025)
“Sequence grammar and dynamics of subcellular translation revealed by APEX-Ribo-Seq.”
BioRxiv. (2025)

Kunitomi H., Khaine A.M., Jamee R., Lancerro M., Rauchendeuri E., Perli S., Sato Y., Iwasaki M., Ruivo P.R., Tomoda K., Shichino Y., Iwasaki S., and Yamanaka S.*:
“eIF4G2-mediated translation initiation of histone modifiers is essential for intestinal stem cell maintenance and differentiation.”
BioRxiv. (2025)
“eIF4G2-mediated translation initiation of histone modifiers is essential for intestinal stem cell maintenance and differentiation.”
BioRxiv. (2025)

Tomuro K. and Iwasaki S.*:
“Advances in ribosome profiling technologies.”
Biochem Soc Trans. 53(3):555–564 (2025)
“Advances in ribosome profiling technologies.”
Biochem Soc Trans. 53(3):555–564 (2025)

Wakigawa T., Mito M., Fang Q., Itoh Y., Shinkai Y., and Iwasaki S.*:
“Mitochondrial translation termination, recycling, reinitiation, and rescue for in-frame and out-of-frame contexts.”
BioRxiv. (2025)
“Mitochondrial translation termination, recycling, reinitiation, and rescue for in-frame and out-of-frame contexts.”
BioRxiv. (2025)

Shichino Y.* and Iwasaki S.:
“Expanding toolkit for RNP granule transcriptomics.”
Polym J. 57(8):873–883 (2025)
“Expanding toolkit for RNP granule transcriptomics.”
Polym J. 57(8):873–883 (2025)

Okubo C.*, Nakamura M., Sato M., Shichino Y., Mito M., Takashima Y., Iwasaki S., and Takahashi K*.:
“Selective translation orchestrates key signaling pathways in primed pluripotency.”
Sci Ad. 11(15):eadq5484 (2025)
BioRxiv. (2024)
Press Release (Kyoto University)
“Selective translation orchestrates key signaling pathways in primed pluripotency.”
Sci Ad. 11(15):eadq5484 (2025)
BioRxiv. (2024)
Press Release (Kyoto University)

Loughran G.*, Andreev D.E., Terenin I.M., Namy O., Mikl M., Yordanova M.M., McManus C.J., Firth A.E., Atkins J.F., Fraser C.S., Ignatova Z., Iwasaki S., Kufel J., Larson O., Leidel S.A., Mankin A.S., Mariotti M., Tanenbaum M.E., Topsirovic I., Vázquez-Laslop N., Viero G., Caliskan N., Chen Y., Clark P.L., Dinman J.D., Farabaugh P.J., Gilbert W.V., Ivanov P., Kieft J.S., Mühlemann O., Sachs M.S., Shatsky I.N., Sonenberg N., Steckelberg A.-L., Willis A.E., Woodside M.T., Valasek L.S.*, Dmitriev S.E.*, Baranov P.V.*.:
“Guidelines for minimal reporting requirements, design and interpretation of experiments involving the use of eukaryotic dual gene expression reporters (MINDR).”
Nat Struct Mol Biol. 32(3):418-430 (2025)
HAL. (2024)
“Guidelines for minimal reporting requirements, design and interpretation of experiments involving the use of eukaryotic dual gene expression reporters (MINDR).”
Nat Struct Mol Biol. 32(3):418-430 (2025)
HAL. (2024)

Fukuchi K.#, Nakashima Y.#, Abe N.#*, Kimura S., Hashiya F., Shichino Y., Liu Y., Ogisu R., Sugiyama S., Kawaguchi D., Inagaki M., Meng Z., Kajihara S., Tada M., Uchida S., Li T.T., Maity R., Kawasaki T, Kimura Y., Iwasaki S., and Abe H.*:
“Internal cap-initiated translation provides efficient protein production from circular mRNA."
Nat Biotechnol. 44(1):120-132 (2026)
Press Release (Nagoya University)
Press Release (RIKEN)
Research Square. (2024)
“Internal cap-initiated translation provides efficient protein production from circular mRNA."
Nat Biotechnol. 44(1):120-132 (2026)
Press Release (Nagoya University)
Press Release (RIKEN)
Research Square. (2024)

Lee M., Wakigawa T., Jia Q., Liu C., Huang R., Huang S., Nagao A., Suzuki T., Tomita K., Iwasaki S., and Takeuchi-Tomita N.*:
“Selection of initiator tRNA and start codon by mammalian mitochondrial initiation factor 3 in leaderless mRNA translation”
Nucleic Acids Res. (2025)
“Selection of initiator tRNA and start codon by mammalian mitochondrial initiation factor 3 in leaderless mRNA translation”
Nucleic Acids Res. (2025)
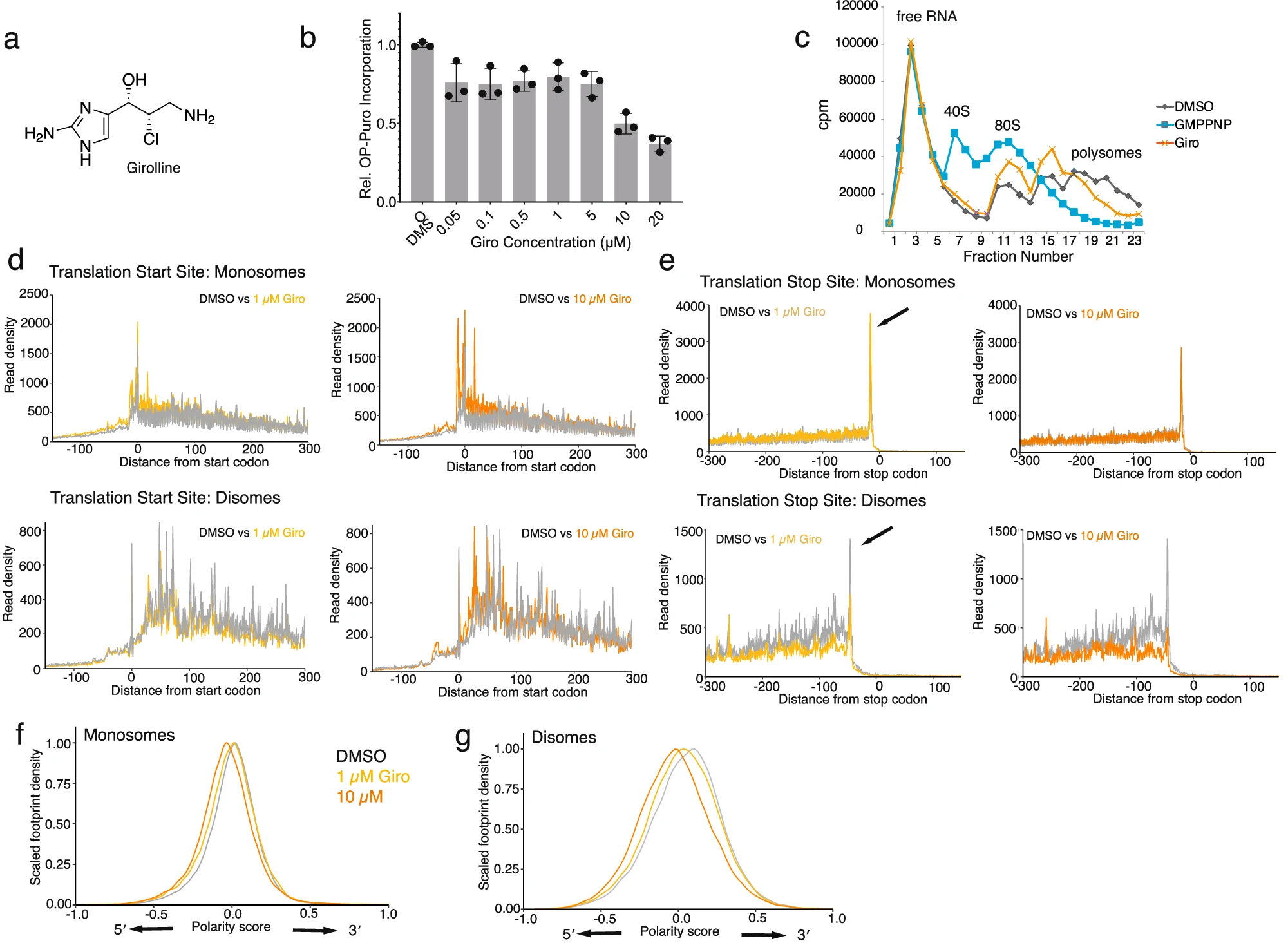
Schneider-Poetsch T.*, Dang Y., Iwasaki W., Arata M., Shichino Y., Mourabit A.A., Moriou C., Romo D., Liu J.O., Ito T., Iwasaki S., and Yoshida M.*:
“Girolline is a sequence context-selective modulator of eIF5A activity”
Nat Commun. 16(1):223 (2025)
Press Release (RIKEN)
Research Square. (2023)
“Girolline is a sequence context-selective modulator of eIF5A activity”
Nat Commun. 16(1):223 (2025)
Press Release (RIKEN)
Research Square. (2023)
Ishibashi K., Shichino Y., Han P., Wakabayashi K., Mito M., Inada T., Kimura S., Iwasaki S., and Mishima Y.*:
“Translation of zinc finger domains induces ribosome collision and Znf598-dependent mRNA decay in zebrafish."
PLoS Biol. 22(12):e3002887 (2024)
Press Release (Kyoto Sangyo University)
Press Release (RIKEN)
BioRxiv. (2024)
Primer
“Translation of zinc finger domains induces ribosome collision and Znf598-dependent mRNA decay in zebrafish."
PLoS Biol. 22(12):e3002887 (2024)
Press Release (Kyoto Sangyo University)
Press Release (RIKEN)
BioRxiv. (2024)
Primer

Iwasaki S.*.:
“New rocaglate derivatives tip the scale against brain tumors.”
ACS Cent Sci. 10(10):1815-1817 (2024)
“New rocaglate derivatives tip the scale against brain tumors.”
ACS Cent Sci. 10(10):1815-1817 (2024)

Matsuura-Suzuki E., Kiyokawa K., Iwasaki S., and Tomari Y.*:
“The requirement of GW182 in miRNA-mediated gene silencing in Drosophila larval development”
EMBO J. 43(23):6161-6179 (2024)
BioRxiv. (2024)
“The requirement of GW182 in miRNA-mediated gene silencing in Drosophila larval development”
EMBO J. 43(23):6161-6179 (2024)
BioRxiv. (2024)
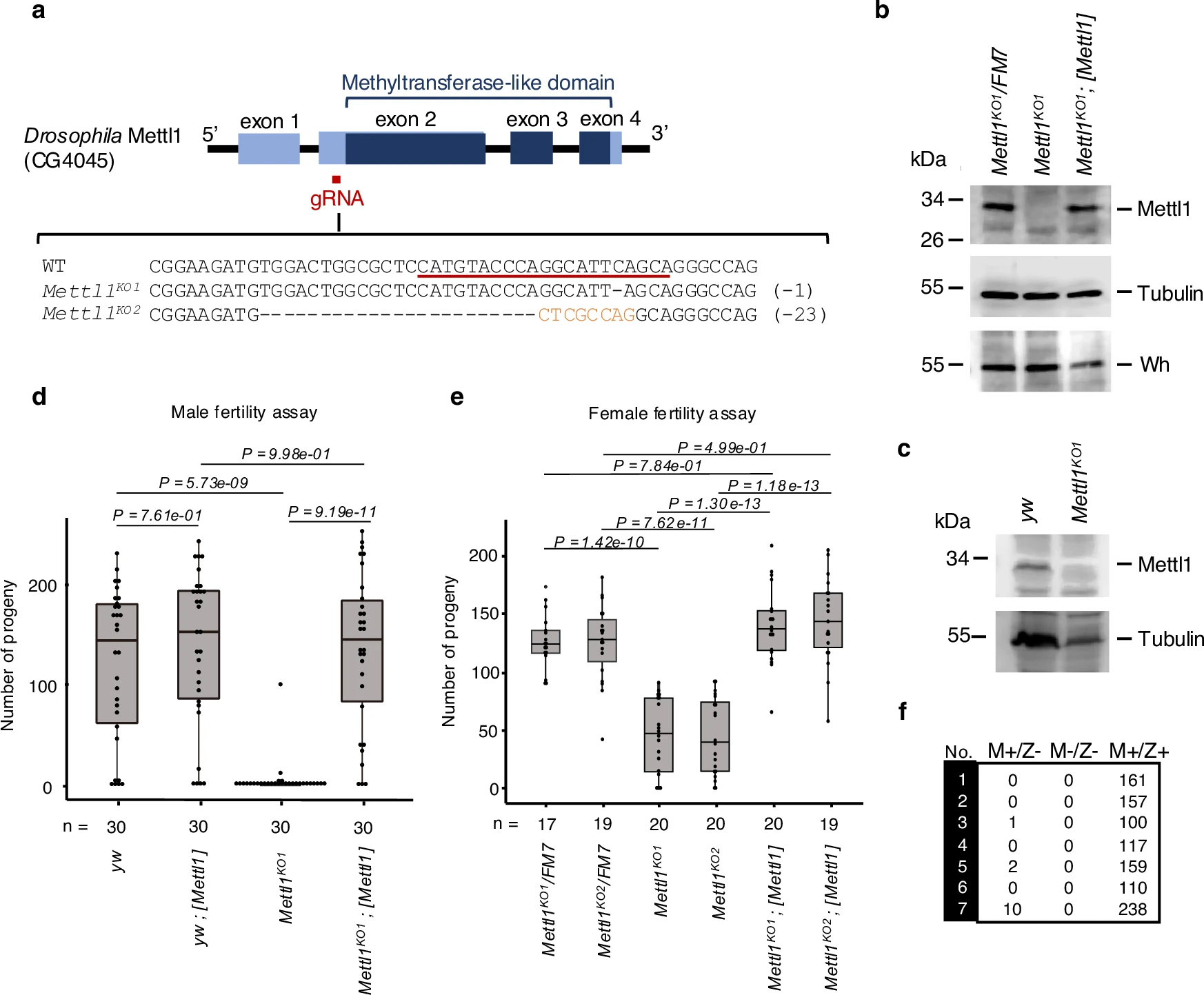
Kaneko S., Miyoshi K., Tomuro K., Terauchi M., Tanaka R., Kondo S., Tani N., Ishiguro K., Toyoda A., Kamikouchi A., Noguchi H., Iwasaki S., and Saito K.*:
“Mettl1-dependent m7G tRNA modification is essential for maintaining spermatogenesis and fertility in Drosophila melanogaster”
Nat Commun. 15(1):8147 (2024)
Press Release (NIG)
BioRxiv. (2023)
“Mettl1-dependent m7G tRNA modification is essential for maintaining spermatogenesis and fertility in Drosophila melanogaster”
Nat Commun. 15(1):8147 (2024)
Press Release (NIG)
BioRxiv. (2023)
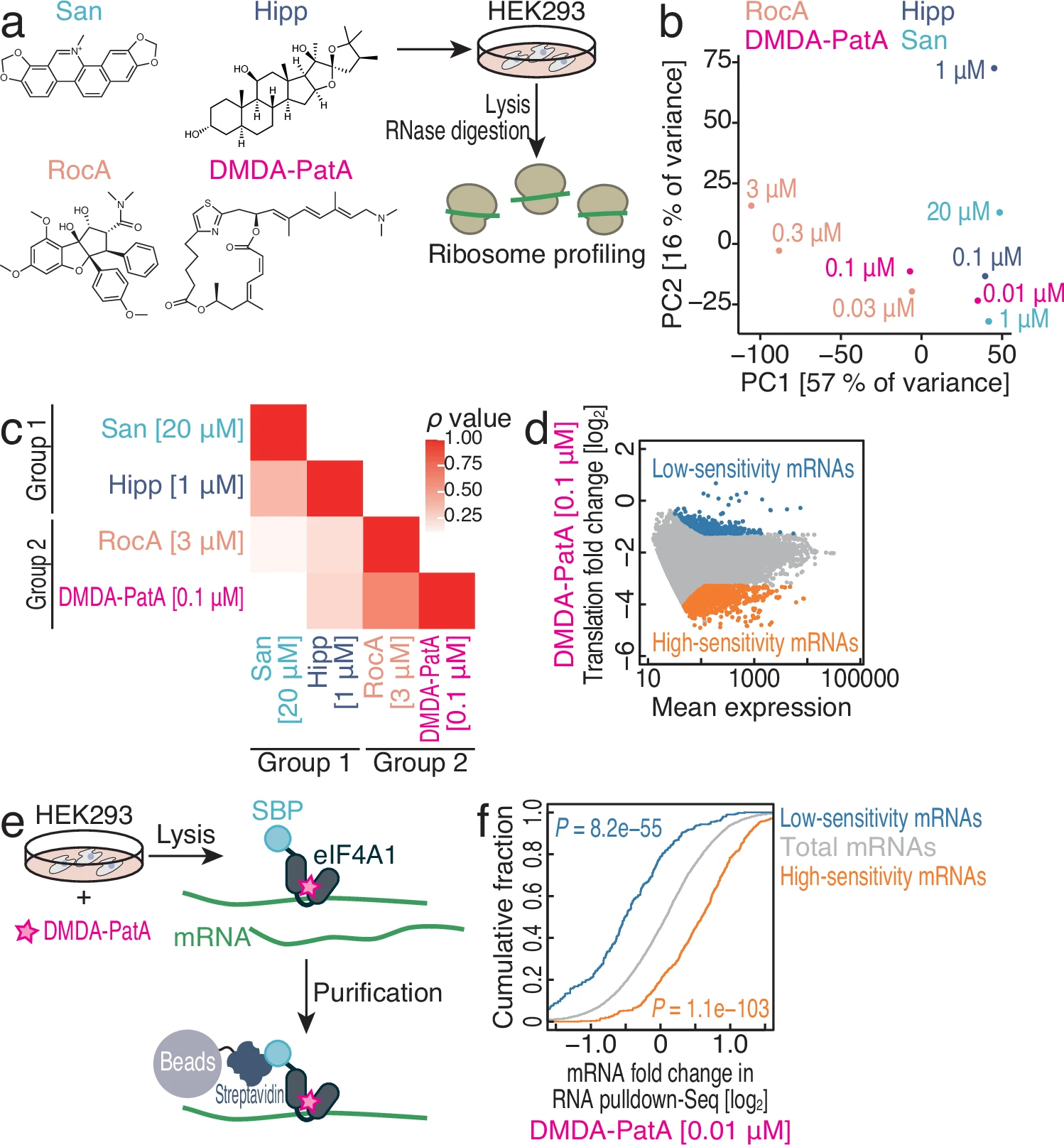
Saito H., Handa Y., Chen M., Schneider-Poetsch T., Shichino Y., Takahashi M., Romo D., Yoshida M., Fürstner A., Ito T., Fukuzawa K., and Iwasaki S.*:
“DMDA-PatA mediates RNA sequence-selective translation repression by anchoring eIF4A and DDX3 to GNG motifs”
Nat Commun. 15(1):7418 (2024)
Addgene
Press Release (RIKEN)
Press Release (Osaka University)
BioRxiv. (2023)
“DMDA-PatA mediates RNA sequence-selective translation repression by anchoring eIF4A and DDX3 to GNG motifs”
Nat Commun. 15(1):7418 (2024)
Addgene
Press Release (RIKEN)
Press Release (Osaka University)
BioRxiv. (2023)
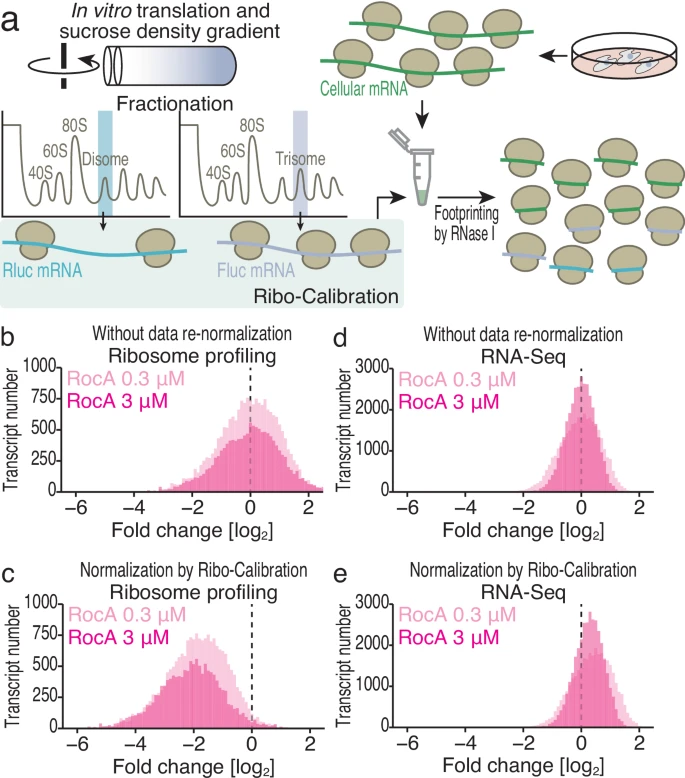
Tomuro K.#, Mito M.#, Toh H., Kawamoto N., Miyake T., Chow S.Y.A., Doi M., Ikeuchi Y., Shichino Y.*, and Iwasaki S.*:
“Calibrated ribosome profiling assesses the dynamics of ribosomal flux on transcripts”
Nat Commun. 15(1):7061 (2024)
Press Release (RIKEN)
Press Release (The University of Tokyo)
Press Release (Nikkei)
BioRxiv. (2024)
“Calibrated ribosome profiling assesses the dynamics of ribosomal flux on transcripts”
Nat Commun. 15(1):7061 (2024)
Press Release (RIKEN)
Press Release (The University of Tokyo)
Press Release (Nikkei)
BioRxiv. (2024)

Tresky R., Miyamoto Y., Nagayoshi Y., Yabuki Y., Araki K., Takahashi Y., Komohara Y., Ge H., Nishiguchi K., Fukuda T., Kaneko H., Maeda N., Matsuura J., Iwasaki S., Sakakida K., Shioda N., Wei F.Y., Tomizawa K.*, Chujo T.*:
“TRMT10A dysfunction perturbs codon translation of initiator methionine and glutamine and impairs brain functions in mice”
Nucleic Acids Res. 52(15):9230-9246 (2024)
Press Release (JST/Kumamoto University)
“TRMT10A dysfunction perturbs codon translation of initiator methionine and glutamine and impairs brain functions in mice”
Nucleic Acids Res. 52(15):9230-9246 (2024)
Press Release (JST/Kumamoto University)

Ichinose T.*, Kondo S., Kanno M., Shichino Y., Mito M., Iwasaki S., and Tanimoto H.*:
“Translational regulation enhances distinction of cell types in the nervous system”
eLife. 12:RP90713 (2024)
BioRxiv. (2024)
“Translational regulation enhances distinction of cell types in the nervous system”
eLife. 12:RP90713 (2024)
BioRxiv. (2024)

Shichino Y.*, Yamaguchi T., Kashiwagi K., Mito M., Takahashi M., Ito T., Ingolia NT., Kuba K., and Iwasaki S.*:
“eIF4A1 enhances LARP1-mediated translational repression during mTORC1 inhibition”
Nat Struct Mol Biol. 31(10):1557-1566 (2024)
Addgene
Press Release (RIKEN)
BioRxiv. (2021)
“eIF4A1 enhances LARP1-mediated translational repression during mTORC1 inhibition”
Nat Struct Mol Biol. 31(10):1557-1566 (2024)
Addgene
Press Release (RIKEN)
BioRxiv. (2021)

Toya H., Okamatsu-Ogura Y., Yokoi S., Kurihara M., Mito M., Iwasaki S., Hirose T., and Nakagawa S*.:
“The essential role of architectural noncoding RNA Neat1 in cold-induced beige adipocyte differentiation in mice”
RNA. 30(8):1011-1024 (2024)
“The essential role of architectural noncoding RNA Neat1 in cold-induced beige adipocyte differentiation in mice”
RNA. 30(8):1011-1024 (2024)
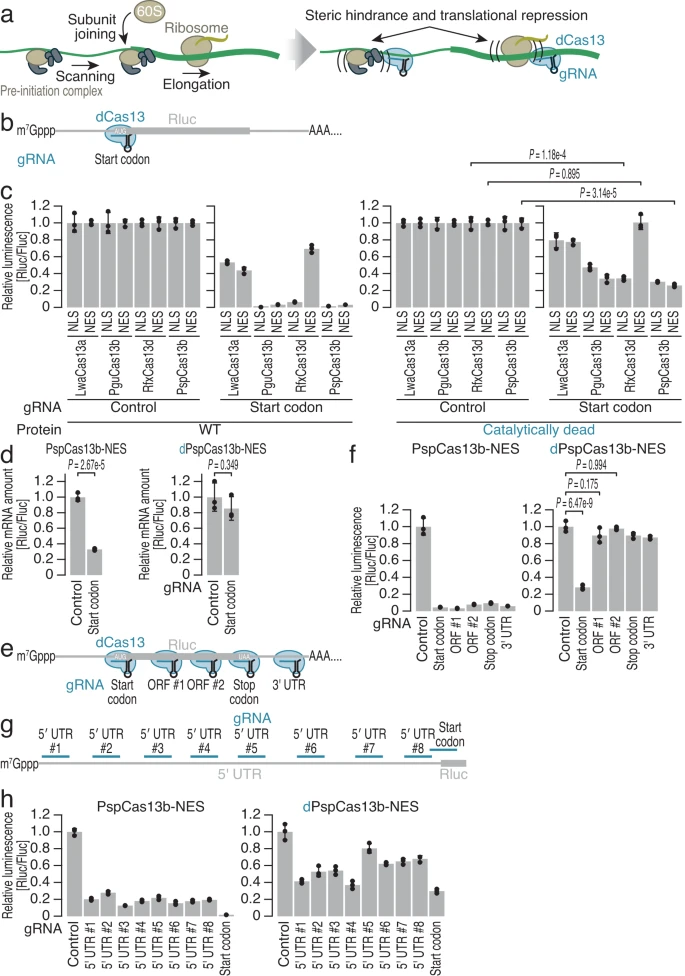
Apostolopoulos A., Kawamoto N., Chow S.Y.A., Tsuiji H., Ikeuchi Y., Shichino Y.*, and Iwasaki S.*:
“dCas13-mediated translational repression for accurate gene silencing in mammalian cells”
Nat Commun. 15(1):2205 (2024)
Addgene
Behind the Paper
Press Release (RIKEN)
Press Release (The University of Tokyo)
BioRxiv. (2023)
“dCas13-mediated translational repression for accurate gene silencing in mammalian cells”
Nat Commun. 15(1):2205 (2024)
Addgene
Behind the Paper
Press Release (RIKEN)
Press Release (The University of Tokyo)
BioRxiv. (2023)

Kumakura N., Singkaravanit-Ogawa S., Gan P., Tsushima A., Ishihama N., Watanabe S., Seo M., Iwasaki S., Narusaka M., Narusaka Y., Takano Y, and Shirasu K.*:
“Guanosine-specific single-stranded ribonuclease effectors of a phytopathogenic fungus potentiate host immune responses”
New Phytol. 242(1):170-191 (2024)
BioRxiv. (2021)
“Guanosine-specific single-stranded ribonuclease effectors of a phytopathogenic fungus potentiate host immune responses”
New Phytol. 242(1):170-191 (2024)
BioRxiv. (2021)
Tanaka M.#, Yokoyama T.#, Saito H.#, Nishimoto M., Tsuda K., Sotta N., Shigematsu H., Shirouzu M., Iwasaki S.*, Ito T.*, and Fujiwara T*.:
“Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.”
Nat Chem Biol. 20(5):605–614 (2024)
Press Release (RIKEN)
Press Release (The University of Tokyo)
“Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.”
Nat Chem Biol. 20(5):605–614 (2024)
Press Release (RIKEN)
Press Release (The University of Tokyo)

Teyssonniere E.M.#, Shichino Y.#, Mito M., Friedrich A., Iwasaki S.*, and Schacherer J.*:
“Translation variation across genetic backgrounds reveals a post-transcriptional buffering signature in yeast.”
Nucleic Acids Res. 52(5):2434-2445 (2024)
BioRxiv. (2023)
“Translation variation across genetic backgrounds reveals a post-transcriptional buffering signature in yeast.”
Nucleic Acids Res. 52(5):2434-2445 (2024)
BioRxiv. (2023)

Wakigawa T.#, Kimura Y.#, Mito M., Tsubaki T., Lee M., Nakamura K., Khan A.H., Saito H., Yamamori T., Yamazaki T., Higashibata A., Tsuboi T., Hirabayashi Y., Takeuchi-Tomita N., Saito T., Higashitani A., Shichino Y., and Iwasaki S.*:
“Gravitational and mechanical forces drive mitochondrial translation.”
BioRxiv. (2024)
“Gravitational and mechanical forces drive mitochondrial translation.”
BioRxiv. (2024)

Matsumoto K.*, Kurokawa R., Takase M., Schneider-Poetsch T., Ling F., Suzuki T., Han P., Wakigawa T., Suzuki M., Tariq M., Ito A., Higashi K., Iwasaki S., Dohmae N., and Yoshida M.*:
“Chemical genetic interaction linking eIF5A hypusination and mitochondrial integrity.”
BioRxiv. (2023)
“Chemical genetic interaction linking eIF5A hypusination and mitochondrial integrity.”
BioRxiv. (2023)

Yoshimoto R.*, Nakayama Y., Nomura I., Yamamoto I., Nakagawa Y., Tanaka S., Kurihara M., Suzuki Y., Kobayashi T., Kozuka-Hata H., Oyama M., Mito M., Iwasaki S., Yamazaki T., Hirose T., Araki K., and Nakagawa S.*:
“4.5SH RNA counteracts deleterious exonization of SINE B1 in mice.”
Mol Cell. S1097-2765(23)00964-4 (2023)
Press Release (Hokkaido University)
Press Release (RIKEN)
Research Square. (2022)
Sneak Peak. (2023)
“4.5SH RNA counteracts deleterious exonization of SINE B1 in mice.”
Mol Cell. S1097-2765(23)00964-4 (2023)
Press Release (Hokkaido University)
Press Release (RIKEN)
Research Square. (2022)
Sneak Peak. (2023)

Zhao X.#, Ma D.#, Ishiguro K.#, Saito H., Akichika S., Matsuzawa I., Mito M. Irie T., Ishibashi K., Wakabayashi K., Sakaguchi Y., Yokoyama T., Mishima Y., Shirouzu M., Iwasaki S., Takeo Suzuki Ta.*, and Suzuki Ts.*:
“Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth”
Cell. 186(25):5517-5535.e24 (2023)
Press Release (The University of Tokyo)
Press Release (RIKEN)
Press Release (Kyoto Sangyo University)
“Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth”
Cell. 186(25):5517-5535.e24 (2023)
Press Release (The University of Tokyo)
Press Release (RIKEN)
Press Release (Kyoto Sangyo University)

Matsuura-Suzuki E.#, Toh H.#, and Iwasaki S.*:
“Human-rabbit hybrid translation system to explore the function of modified ribosomes”
Bio protoc. 13(13):e4714. (2023)
“Human-rabbit hybrid translation system to explore the function of modified ribosomes”
Bio protoc. 13(13):e4714. (2023)
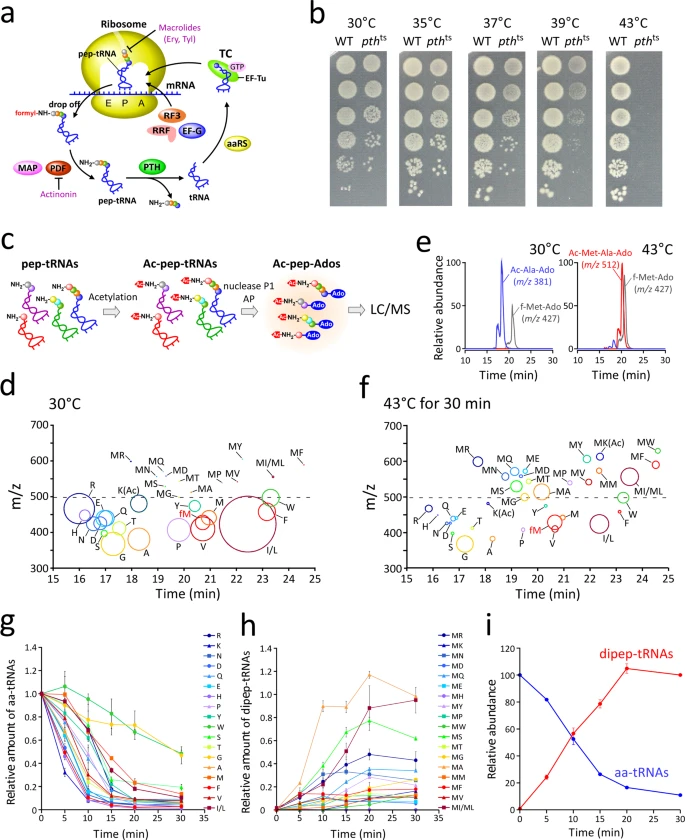
Nagao A.*, Nakanishi Y., Yamaguchi Y., Mishina Y., Karoji M., Toya T., Fujita T., Iwasaki S., Miyauchi K., Sakaguchi Y., and Suzuki T.*:
“Quality control of protein synthesis in the early elongation stage”
Nat Commun. 14(1):2704 (2023)
Press Release (The University of Tokyo)
Press Release (Nikkei)
“Quality control of protein synthesis in the early elongation stage”
Nat Commun. 14(1):2704 (2023)
Press Release (The University of Tokyo)
Press Release (Nikkei)

Kito Y.#, Matsumoto A.#*, Ichihara K., Shiraishi C., Tang R., Hatano A., Matsumoto M., Han P., Iwasaki S., and Nakayama KI.*:
“The ASC-1 complex promotes translation initiation by scanning ribosomes”
EMBO J. 42(12):e112869 (2023)
Press Release (RIKEN)
Press Release (Kyoto University)
“The ASC-1 complex promotes translation initiation by scanning ribosomes”
EMBO J. 42(12):e112869 (2023)
Press Release (RIKEN)
Press Release (Kyoto University)
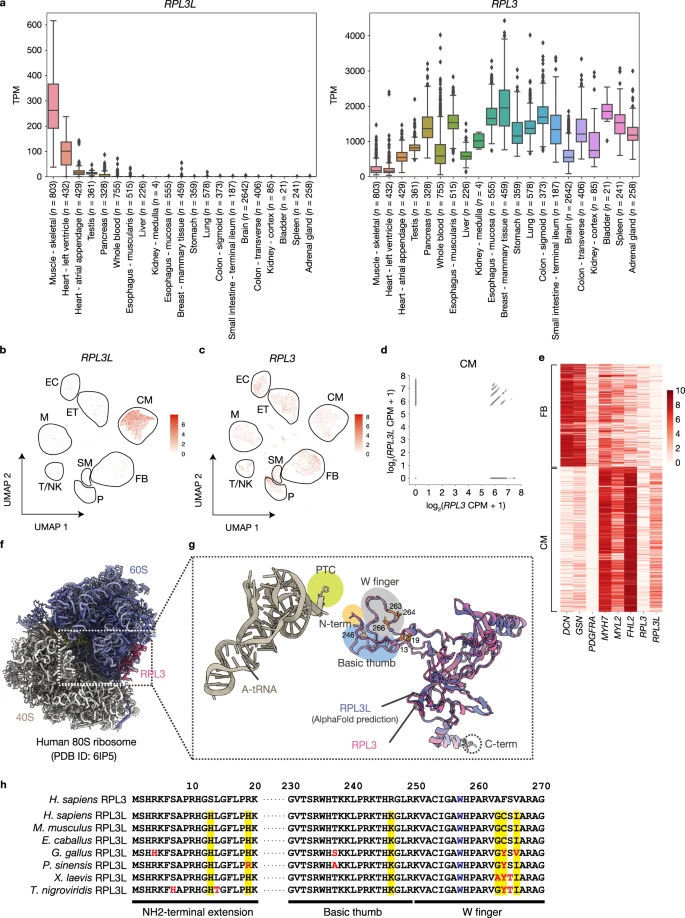
Shiraishi C.#, Matsumoto A.#*, Ichihara K., Yamamoto T., Yokoyama T., Mizoo T., Hatano A., Matsumoto M., Tanaka Y., Matsuura-Suzuki E., Iwasaki S., Matsushima S., Tsutsui H., and Nakayama KI.*:
“RPL3L-containing ribosomes determine translation elongation dynamics required for cardiac function”
Nat Commun. 14(1):2131 (2023)
Press Release (RIKEN)
Press Release (Kyoto University)
“RPL3L-containing ribosomes determine translation elongation dynamics required for cardiac function”
Nat Commun. 14(1):2131 (2023)
Press Release (RIKEN)
Press Release (Kyoto University)

Miyake T.#, Inoue Y.#, Shao X., Seta T., Aoki Y., Nguyen Pham KT., Shichino Y., Sasaki J., Sasaki T., Ikawa M., Yamaguchi Y., Okamura H., Iwasaki S., and Doi M.*:
“Minimal upstream ORF of Per2 mediates phase fitness of the circadian clock to day/night physiological body temperature rhythm”
Cell Rep. 42(3):112157 (2023)
Press Release (Kyoto University)
“Minimal upstream ORF of Per2 mediates phase fitness of the circadian clock to day/night physiological body temperature rhythm”
Cell Rep. 42(3):112157 (2023)
Press Release (Kyoto University)
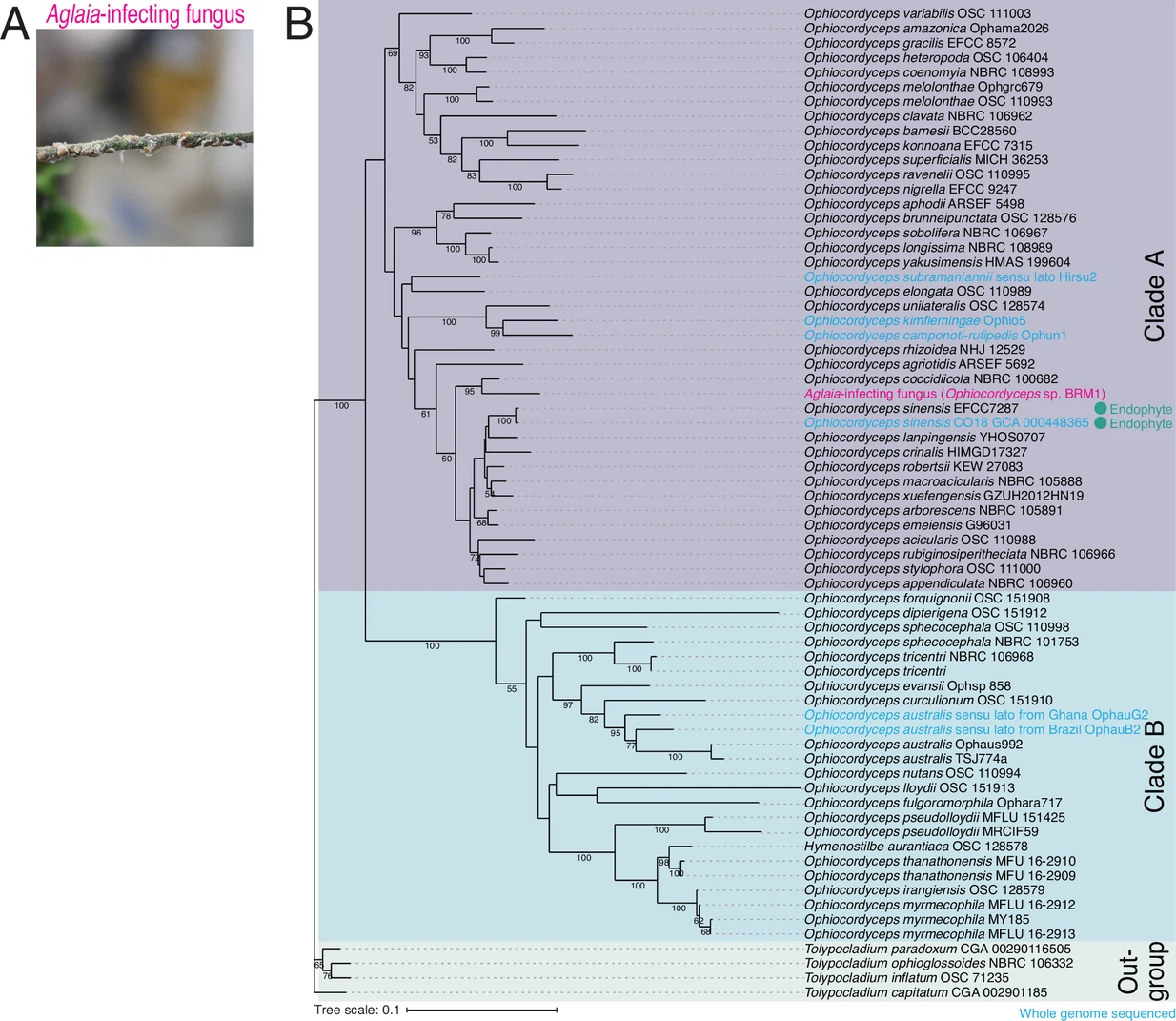
Chen M.#, Kumakura N.#, Muller R., Shichino Y., Nishimoto M., Mito M., Gan P., Ingolia NT., Shirasu K., Ito T., and Iwasaki S.*:
“A parasitic fungus employs mutated eIF4A to survive on rocaglate-synthesizing Aglaia plants”
eLife. 12:e81302. (2023)
BioRxiv. (2022)
Press Release (RIKEN)
科学道2023 (RIKEN)
RIKEN Research Highlight (RIKEN)
“A parasitic fungus employs mutated eIF4A to survive on rocaglate-synthesizing Aglaia plants”
eLife. 12:e81302. (2023)
BioRxiv. (2022)
Press Release (RIKEN)
科学道2023 (RIKEN)
RIKEN Research Highlight (RIKEN)

Yamashita A., Shichino Y., Fujii K., Koshidaka Y., Adachi M., Sasagawa E., Mito M., Nakagawa S., Iwasaki S., Takao K., and Shiina N*.:
“ILF3 prion-like domain regulates gene expression and fear memory under chronic stress”
iScience. 26(3):106229. (2023)
Press Release (RIKEN)
Press Release (NIBB)
“ILF3 prion-like domain regulates gene expression and fear memory under chronic stress”
iScience. 26(3):106229. (2023)
Press Release (RIKEN)
Press Release (NIBB)

Saito H., Osaki T., Ikeuchi Y., and Iwasaki S.*:
“High-throughput assessment of mitochondrial protein synthesis in mammalian cells using mito-FUNCAT FACS”
Bio protoc. 13(3):e4602. (2023)
Journal Cover
“High-throughput assessment of mitochondrial protein synthesis in mammalian cells using mito-FUNCAT FACS”
Bio protoc. 13(3):e4602. (2023)
Journal Cover

Mito M.#, Shichino Y.#, and Iwasaki S.*:
“Thor-Ribo-Seq: ribosome profiling tailored for low input with RNA-dependent RNA amplification”
BioRxiv. (2023)
“Thor-Ribo-Seq: ribosome profiling tailored for low input with RNA-dependent RNA amplification”
BioRxiv. (2023)
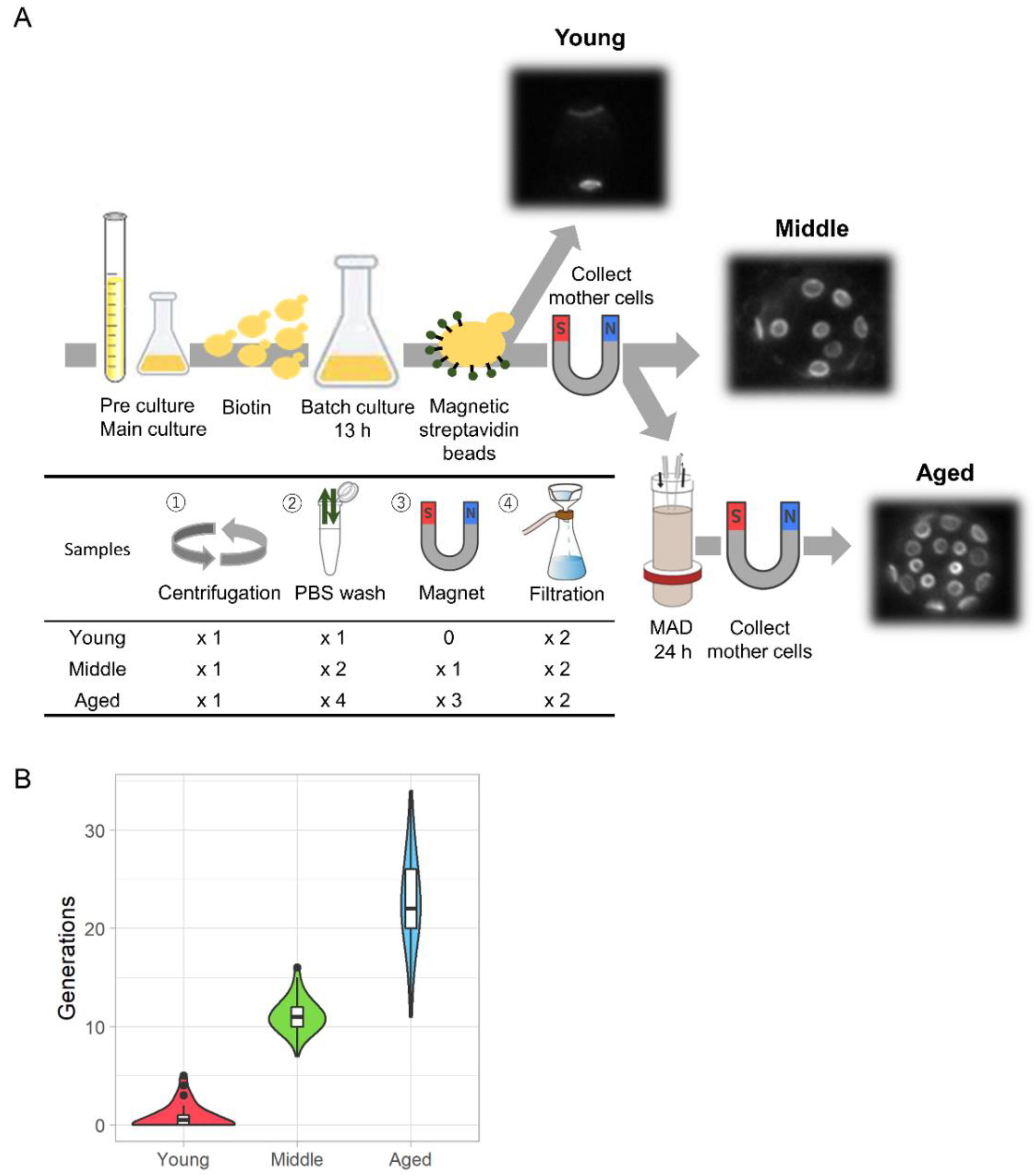
Zhao T.#, Chida A.#, Shichino Y.#, Choi D., Mizunuma M., Iwasaki S., and Ohya Y.*:
“Multifarious translational regulation during replicative aging in yeast”
J Fungi (Basal). 8(9):938. (2022)
“Multifarious translational regulation during replicative aging in yeast”
J Fungi (Basal). 8(9):938. (2022)
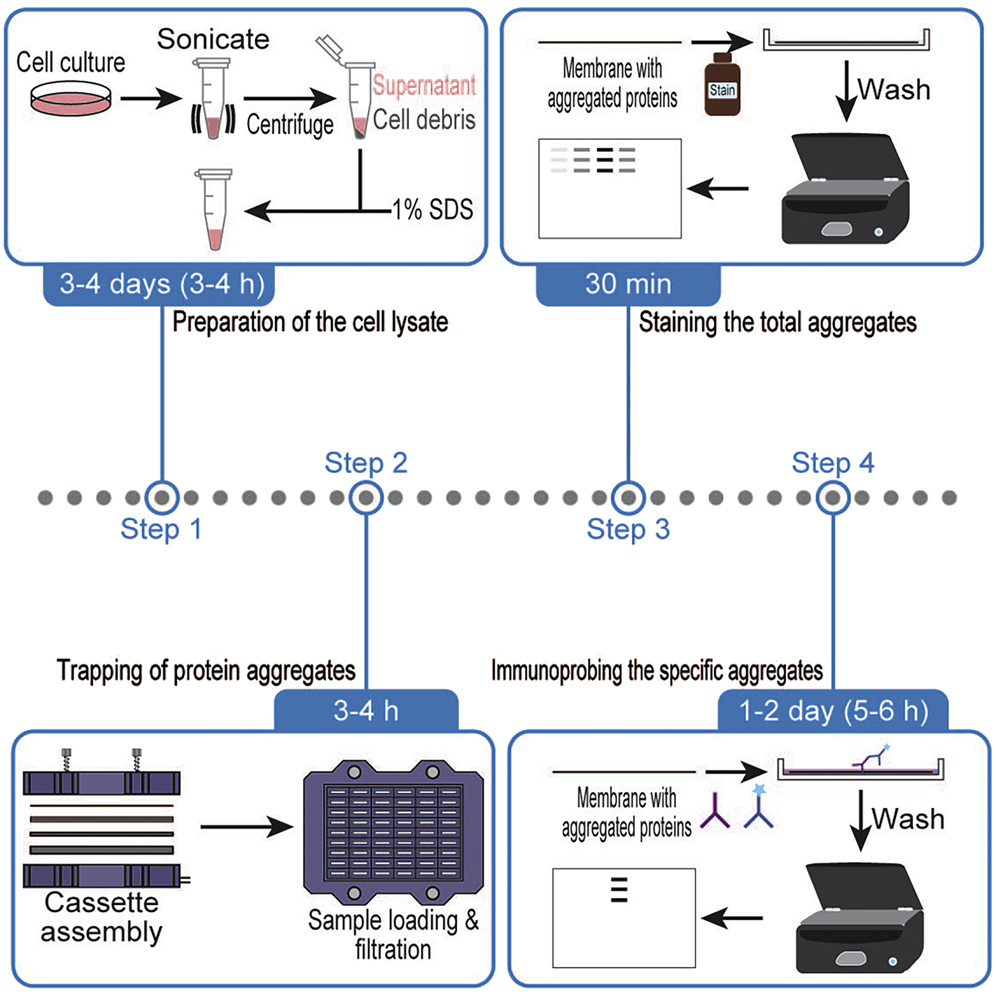
Chhipi-Shrestha JK., Yoshida M.*, and Iwasaki S.*:
“Filter trapping protocol to detect aggregated proteins in human cell lines”
STAR Protoc. 3(3):101571. (2022)
“Filter trapping protocol to detect aggregated proteins in human cell lines”
STAR Protoc. 3(3):101571. (2022)
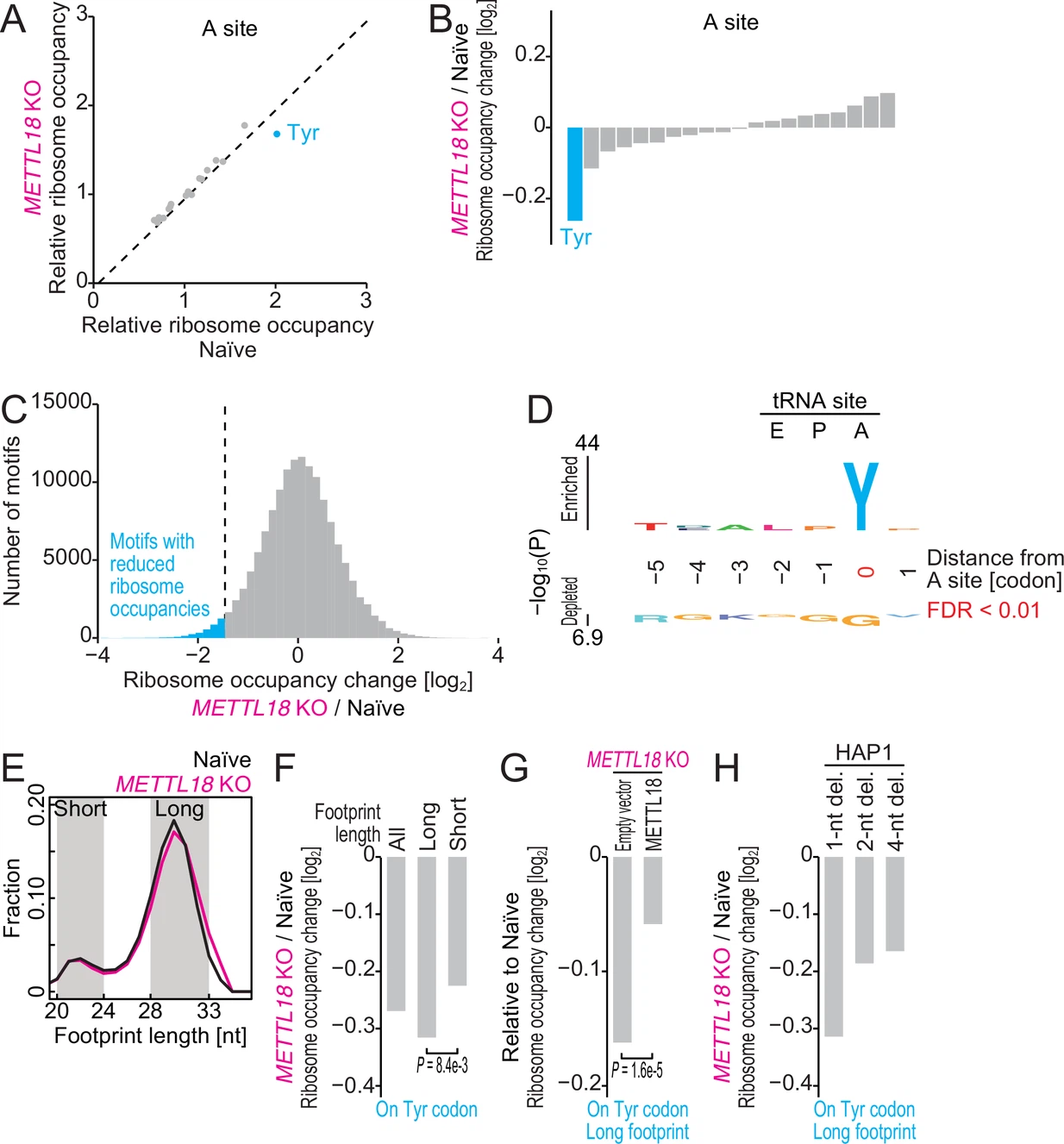
Matsuura-Suzuki E.#, Shimazu T.#*, Takahashi M., Kotoshiba K., Suzuki T., Kashiwagi K., Sohtome Y., Akakabe M., Sodeoka M., Dohmae N., Ito T.*, Shinkai Y.*, and Iwasaki S.*:
“METTL18-mediated histidine methylation on RPL3 modulates translation elongation for proteostasis maintenance”
eLife. 11:e72780. (2022)
Addgene
BioRxiv. (2021)
Press Release (RIKEN)
“METTL18-mediated histidine methylation on RPL3 modulates translation elongation for proteostasis maintenance”
eLife. 11:e72780. (2022)
Addgene
BioRxiv. (2021)
Press Release (RIKEN)

Yamada A., Toya H., Tanahashi M., Kurihara M., Mito M., Iwasaki S., Kurosaka S., Takumi T., Fox A., Kawamura Y., Miura K., and Nakagawa S.*:
“Species-specific formation of paraspeckles in intestinal epithelium revealed by characterization of NEAT1 in naked mole-rat”
RNA. 28(8):1128-1143. (2022)
RNA Journal Twitter (2022)
BioRxiv. (2022)
“Species-specific formation of paraspeckles in intestinal epithelium revealed by characterization of NEAT1 in naked mole-rat”
RNA. 28(8):1128-1143. (2022)
RNA Journal Twitter (2022)
BioRxiv. (2022)

Shichino Y.* and Iwasaki S.*:
“Compounds for selective translational inhibition”
Curr Opin Chem Biol. 69:102158. (2022)
“Compounds for selective translational inhibition”
Curr Opin Chem Biol. 69:102158. (2022)
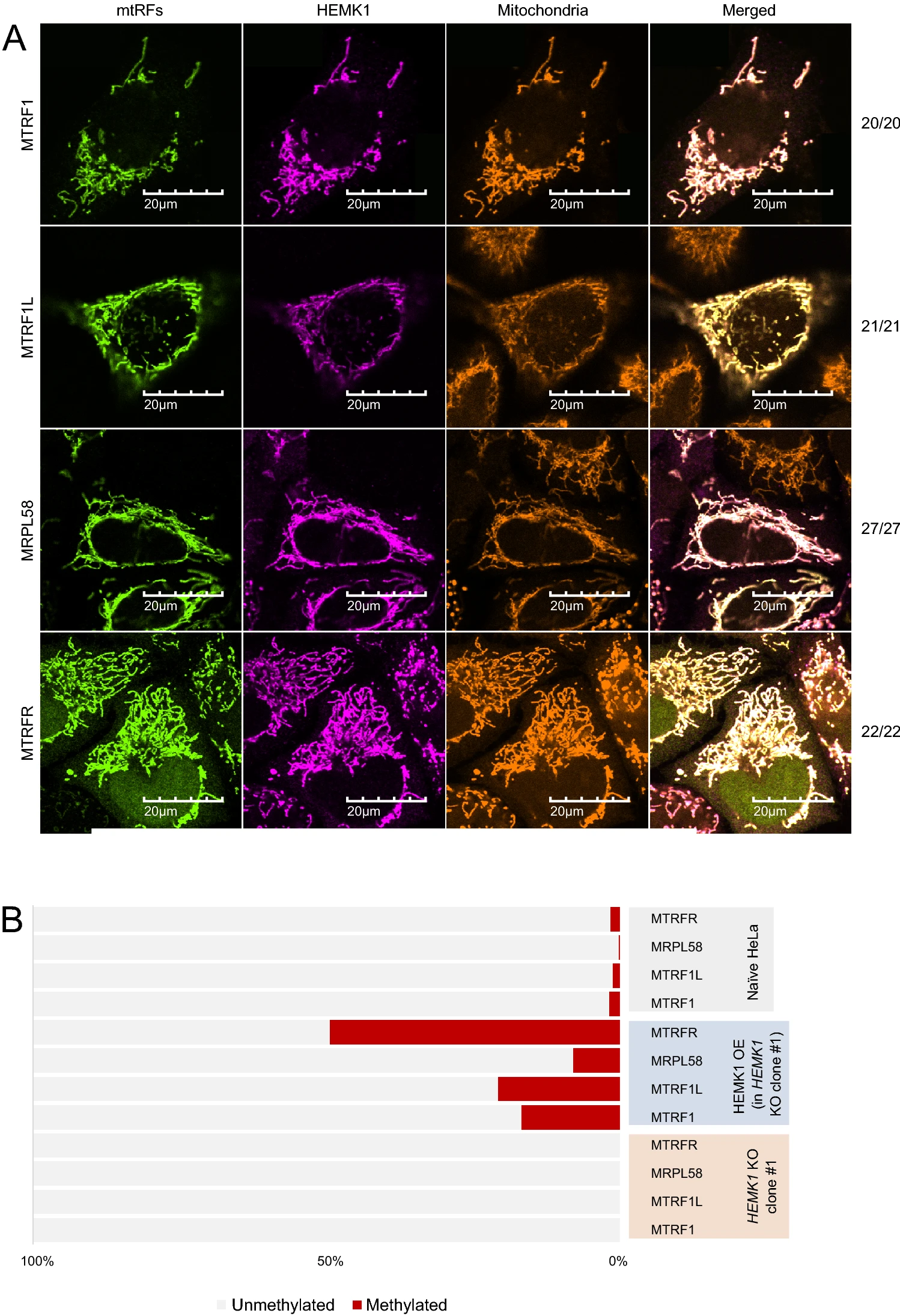
Fang Q., Kimura Y., Shimazu T., Suzuki T., Dohmae N., Iwasaki S., and Shinkai Y.*:
“HEMK1 methylates glutamine residue of the GGQ motif of mitochondrial release factors”
Sci Rep. 12(1):4104. (2022)
Research Square. (2021)
“HEMK1 methylates glutamine residue of the GGQ motif of mitochondrial release factors”
Sci Rep. 12(1):4104. (2022)
Research Square. (2021)

Kimura Y.#, Saito H.#, Osaki T., Ikegami Y., Wakigawa T., Ikeuchi Y., and Iwasaki S.*:
“Mito-FUNCAT-FACS reveals cellular heterogeneity in mitochondrial translation”
RNA. 28(6):895-904. (2022)
RNA Journal Twitter (2022)
BioRxiv. (2022)
“Mito-FUNCAT-FACS reveals cellular heterogeneity in mitochondrial translation”
RNA. 28(6):895-904. (2022)
RNA Journal Twitter (2022)
BioRxiv. (2022)
Apostolopoulos A. and Iwasaki S.*:
“Into the matrix: current methods for mitochondrial translation studies”
J Biochem. 171(4):379-387. (2022)
“Into the matrix: current methods for mitochondrial translation studies”
J Biochem. 171(4):379-387. (2022)
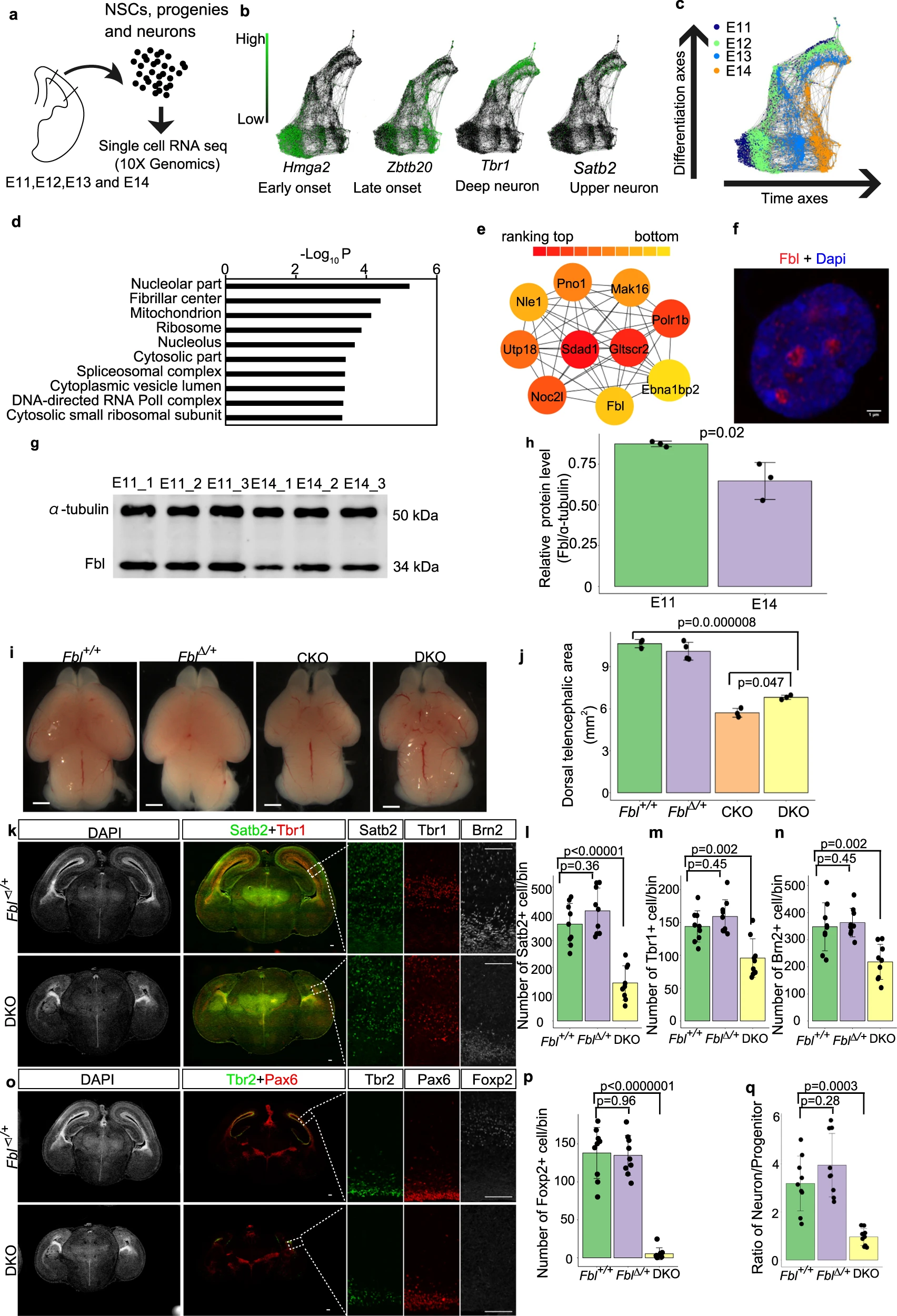
Wu Q., Shichino Y., Abe T., Suetsugu T., Omori A., Kiyonari H., Iwasaki S., and Matsuzaki F.*:
“Selective translation of epigenetic modifiers affects the temporal pattern and differentiation of neural stem cells”
Nat Commun. 13(1):470. (2022)
BioRxiv. (2020)
“Selective translation of epigenetic modifiers affects the temporal pattern and differentiation of neural stem cells”
Nat Commun. 13(1):470. (2022)
BioRxiv. (2020)

Mishima Y.*, Han P., Ishibashi K., Kimura S., and Iwasaki S.:
“Ribosome slowdown triggers codon-mediated mRNA decay independently of ribosome quality control”
EMBO J. 41(5):e109256. (2022)
BioRxiv. (2021)
Press Release (RIKEN)
Press Release (Kyoto Sangyo University)
Research Results (AMED)
“Ribosome slowdown triggers codon-mediated mRNA decay independently of ribosome quality control”
EMBO J. 41(5):e109256. (2022)
BioRxiv. (2021)
Press Release (RIKEN)
Press Release (Kyoto Sangyo University)
Research Results (AMED)

Fujita T., Yokoyama T., Shirouzu M., Taguchi H., Ito T., and Iwasaki S.*:
“The landscape of translational stall sites in bacteria revealed by monosome and disome profiling”
RNA. 28(3):290-302 (2022)
RNA Journal Twitter (2022)
“The landscape of translational stall sites in bacteria revealed by monosome and disome profiling”
RNA. 28(3):290-302 (2022)
RNA Journal Twitter (2022)
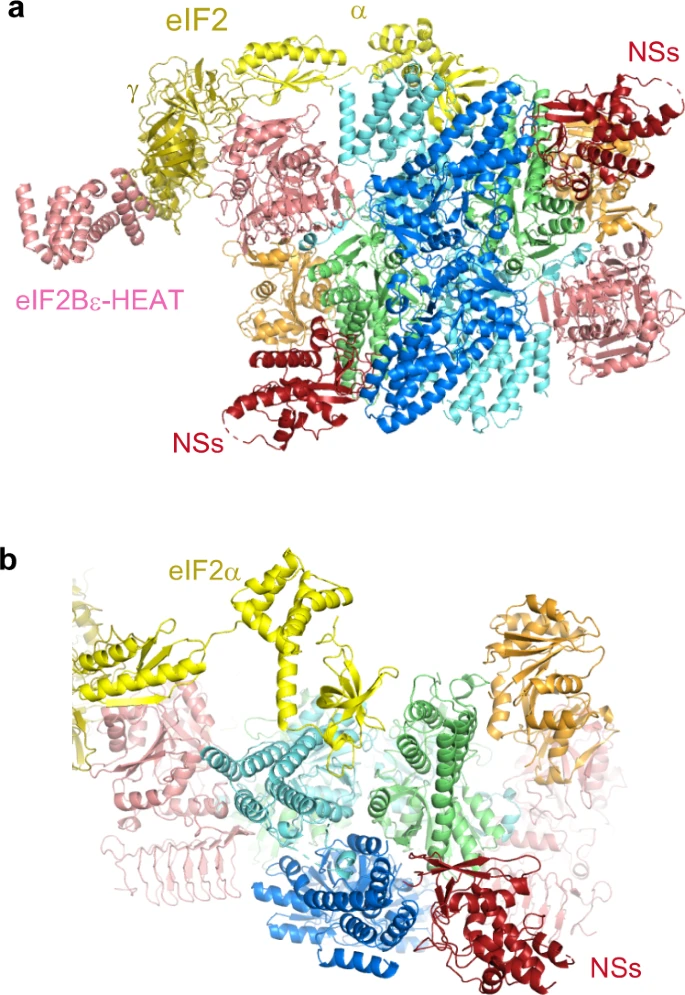
Kashiwagi K.#, Shichino Y.#, Osaki T.#, Sakamoto A., Nishimoto M., Takahashi M., Mito M., Weber F., Ikeuchi Y.*, Iwasaki S.*, and Ito T.*:
“eIF2B-capturing viral protein NSs suppresses the integrated stress response”
Nat Commun. 12(1):7102. (2021)
BioRxiv. (2021)
Press Release (RIKEN)
Press Release (The University of Tokyo)
Research Results (AMED)
RIKEN Research Highlight (RIKEN)
“eIF2B-capturing viral protein NSs suppresses the integrated stress response”
Nat Commun. 12(1):7102. (2021)
BioRxiv. (2021)
Press Release (RIKEN)
Press Release (The University of Tokyo)
Research Results (AMED)
RIKEN Research Highlight (RIKEN)

Chadani Y.*, Sugata N., Niwa T., Ito Y., Iwasaki S., and Taguchi H.*:
“Nascent polypeptide within the exit tunnel stabilizes the ribosome to counteract risky translation”
EMBO J. 40(23):e108299. (2021)
BioRxiv. (2021)
Press Release (RIKEN)
Press Release (Tokyo Institute of Technology)
“Nascent polypeptide within the exit tunnel stabilizes the ribosome to counteract risky translation”
EMBO J. 40(23):e108299. (2021)
BioRxiv. (2021)
Press Release (RIKEN)
Press Release (Tokyo Institute of Technology)

Chhipi-Shrestha JK., Schneider-Poetsch T., Suzuki T., Mito M., Kha K., Dohmae N., Iwasaki S.*, and Yoshida M.*:
“Splicing modulators elicit global translational repression by condensate-prone proteins translated from introns”
Cell Chem Biol. 29(2):259-275.e10. (2022)
BioRxiv. (2020)
Press Release (RIKEN)
Press Release (The University of Tokyo)
Press Release (Nikkei)
“Splicing modulators elicit global translational repression by condensate-prone proteins translated from introns”
Cell Chem Biol. 29(2):259-275.e10. (2022)
BioRxiv. (2020)
Press Release (RIKEN)
Press Release (The University of Tokyo)
Press Release (Nikkei)
Ichihara K., Matsumoto A.*, Nishida H., Kito Y., Shimizu H., Shichino Y., Iwasaki S., Imami K., Ishihama Y., and Nakayama KI.*:
“Combinatorial analysis of translation dynamics reveals eIF2 dependence of translation initiation at near-cognate codons”
Nucleic Acids Res. 49(13):7298-7317. (2021)
Press Release (RIKEN)
Press Release (Kyushu University)
“Combinatorial analysis of translation dynamics reveals eIF2 dependence of translation initiation at near-cognate codons”
Nucleic Acids Res. 49(13):7298-7317. (2021)
Press Release (RIKEN)
Press Release (Kyushu University)

Iwakawa HO.*, Lam Y.W.A., Mine A., Fujita T., Kiyokawa K., Yoshikawa M., Takeda A., Iwasaki S., and Tomari Y.:
“Ribosome stalling caused by the Argonaute-miRNA-SGS3 complex regulates production of secondary siRNA biogenesis in plants”
Cell Rep. 35(13):109300. (2021)
BioRxiv. (2020)
Press Release (RIKEN)
Press Release (The University of Tokyo)
“Ribosome stalling caused by the Argonaute-miRNA-SGS3 complex regulates production of secondary siRNA biogenesis in plants”
Cell Rep. 35(13):109300. (2021)
BioRxiv. (2020)
Press Release (RIKEN)
Press Release (The University of Tokyo)
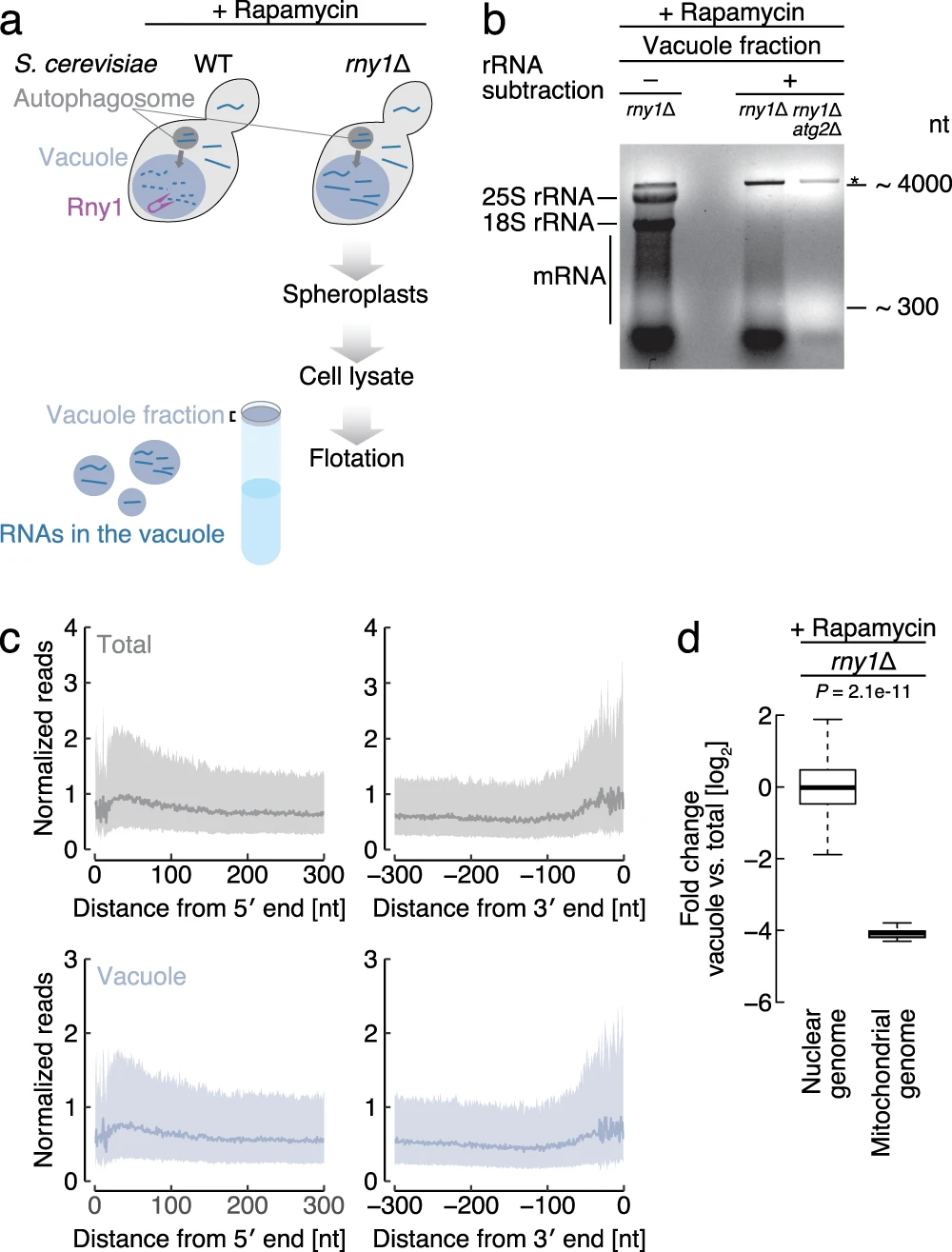
Makino S., Kawamata T., Iwasaki S.*, and Ohsumi Y.*:
“Selectivity of mRNA degradation by autophagy in yeast”
Nat Commun. 12(1):2316. (2021)
Press Release (RIKEN)
Press Release (Tokyo Institute of Technology)
Editors’ Highlights
Recommended in Faculty Opinions
“Selectivity of mRNA degradation by autophagy in yeast”
Nat Commun. 12(1):2316. (2021)
Press Release (RIKEN)
Press Release (Tokyo Institute of Technology)
Editors’ Highlights
Recommended in Faculty Opinions

Yoshimoto R., Chhipi-Shrestha JK., Schneider-Poetsch T., Furuno M., Burroughs MA., Noma S., Suzuki H., Hayashizaki Y., Mayeda A., Nakagawa S., Kaida D., Iwasaki S.*, and Yoshida M.*:
“Spliceostatin A interaction with SF3B1 limits U1 snRNP availability and causes premature cleavage and polyadenylation”
Cell Chem Biol. 28(9):1356-1365.e4. (2021)
Press Release
“Spliceostatin A interaction with SF3B1 limits U1 snRNP availability and causes premature cleavage and polyadenylation”
Cell Chem Biol. 28(9):1356-1365.e4. (2021)
Press Release
Higashi H., Kato Y., Fujita T., Iwasaki S., Nakamura M., Nishimura Y., Takenaka M., and Shikanai T.*:
“The pentatricopeptide repeat protein PGR3 is required for the translation of petL and ndhG by binding their 5'UTRs”
Plant Cell Physiol. 62(7):1146–1155. (2021)
“The pentatricopeptide repeat protein PGR3 is required for the translation of petL and ndhG by binding their 5'UTRs”
Plant Cell Physiol. 62(7):1146–1155. (2021)

Chen M., Asanuma M., Takahashi M., Shichino Y., Mito M., Fujiwara K., Saito H., Floor SN., Ingolia NT., Sodeoka M., Dodo K., Ito T., and Iwasaki S.*:
“Dual targeting of DDX3 and eIF4A by the translation inhibitor rocaglamide A”
Cell Chem Biol. 28(4):475-486.e8. (2021)
Addgene
Press Release
Journal Cover
Preview
“Dual targeting of DDX3 and eIF4A by the translation inhibitor rocaglamide A”
Cell Chem Biol. 28(4):475-486.e8. (2021)
Addgene
Press Release
Journal Cover
Preview
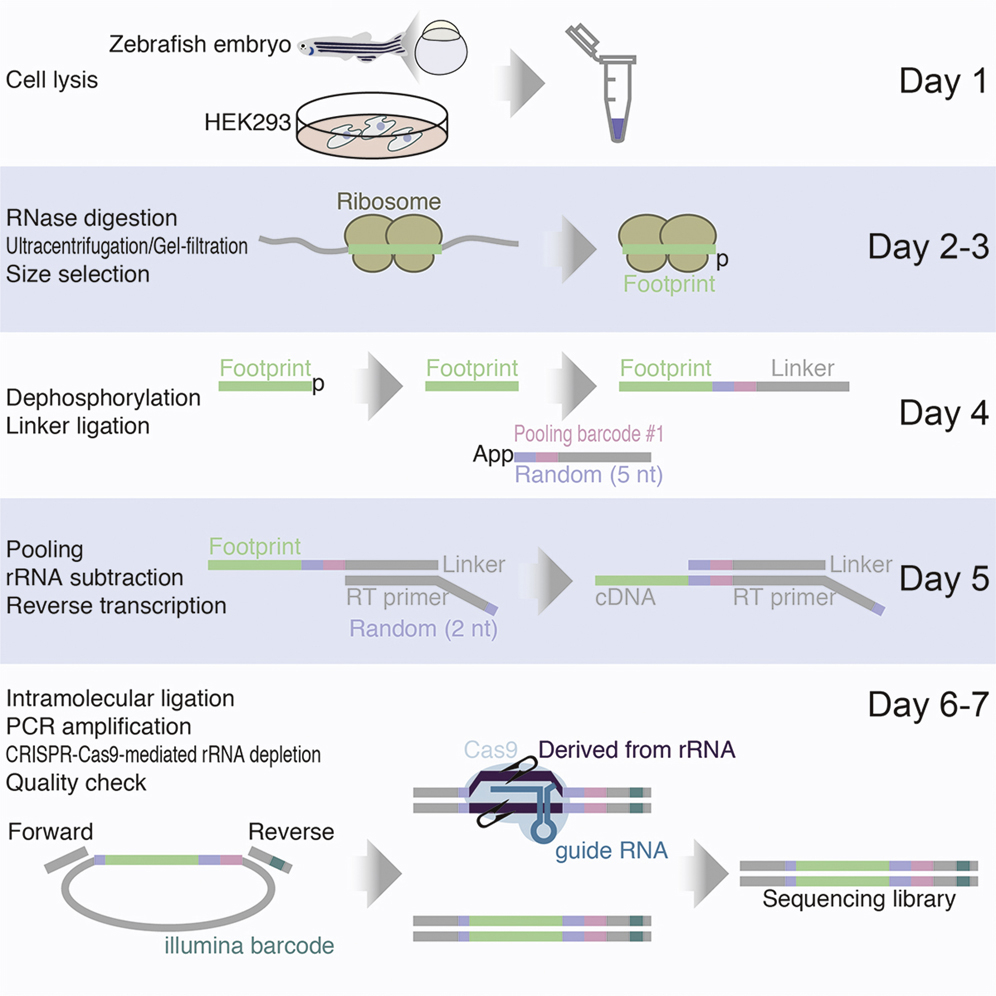
Mito M., Mishima Y., and Iwasaki S.*:
“Protocol for disome profiling to survey ribosome collision in humans and zebrafish”
STAR Protoc. 1(3):100168. (2020)
“Protocol for disome profiling to survey ribosome collision in humans and zebrafish”
STAR Protoc. 1(3):100168. (2020)
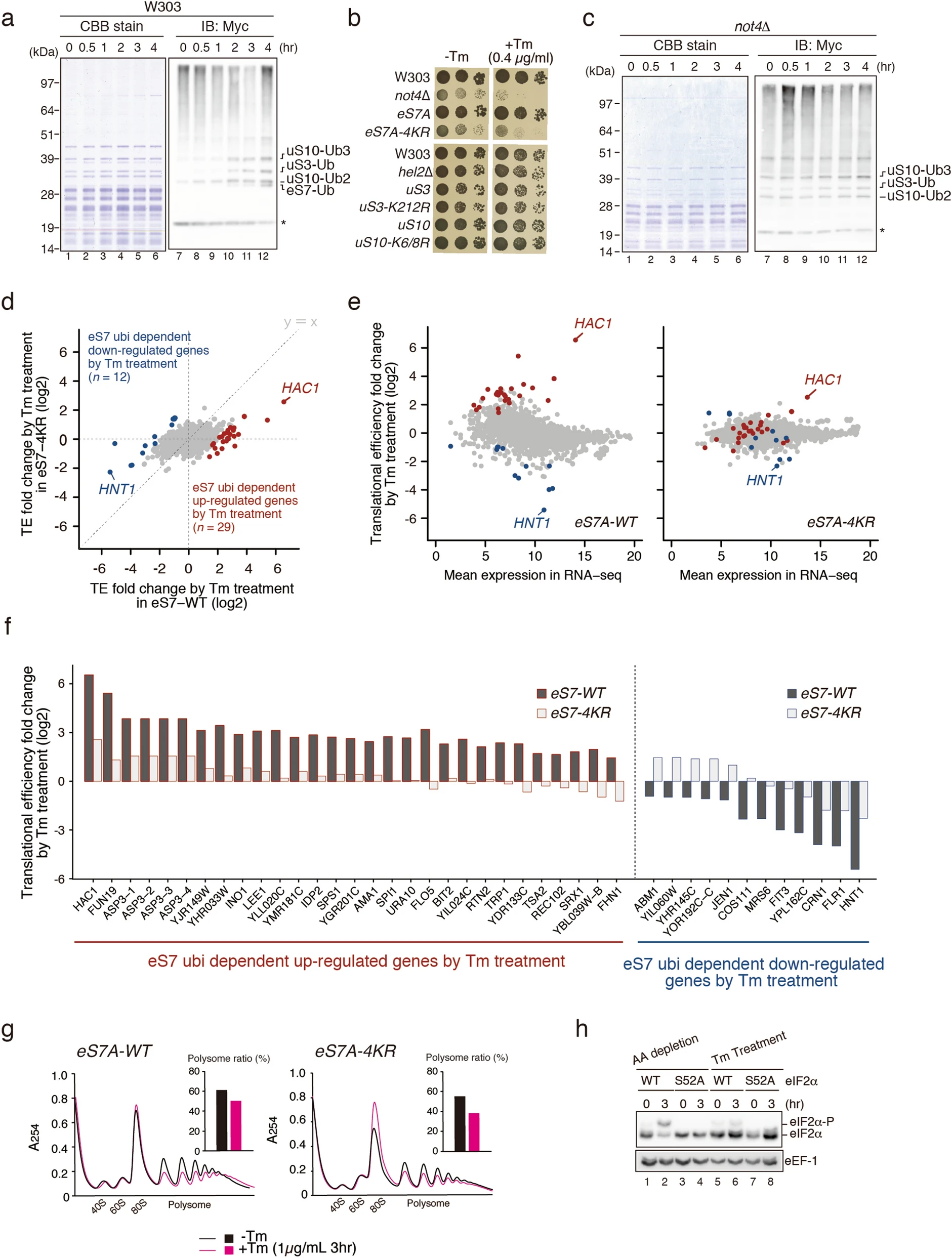
Matsuki Y., Matsuo y., Nakano Y., Iwasaki S., Yoko H., Udagawa T., Li S., Saeki Y., Yoshihisa T., Tanaka K., Ingolia NT., and Inada T.*:
“Ribosomal protein S7 ubiquitination during ER stress in yeast is associated with selective mRNA translation and stress outcome”
Sci Rep. 110(1):19669. (2020)
BioRxiv. (2020)
“Ribosomal protein S7 ubiquitination during ER stress in yeast is associated with selective mRNA translation and stress outcome”
Sci Rep. 110(1):19669. (2020)
BioRxiv. (2020)
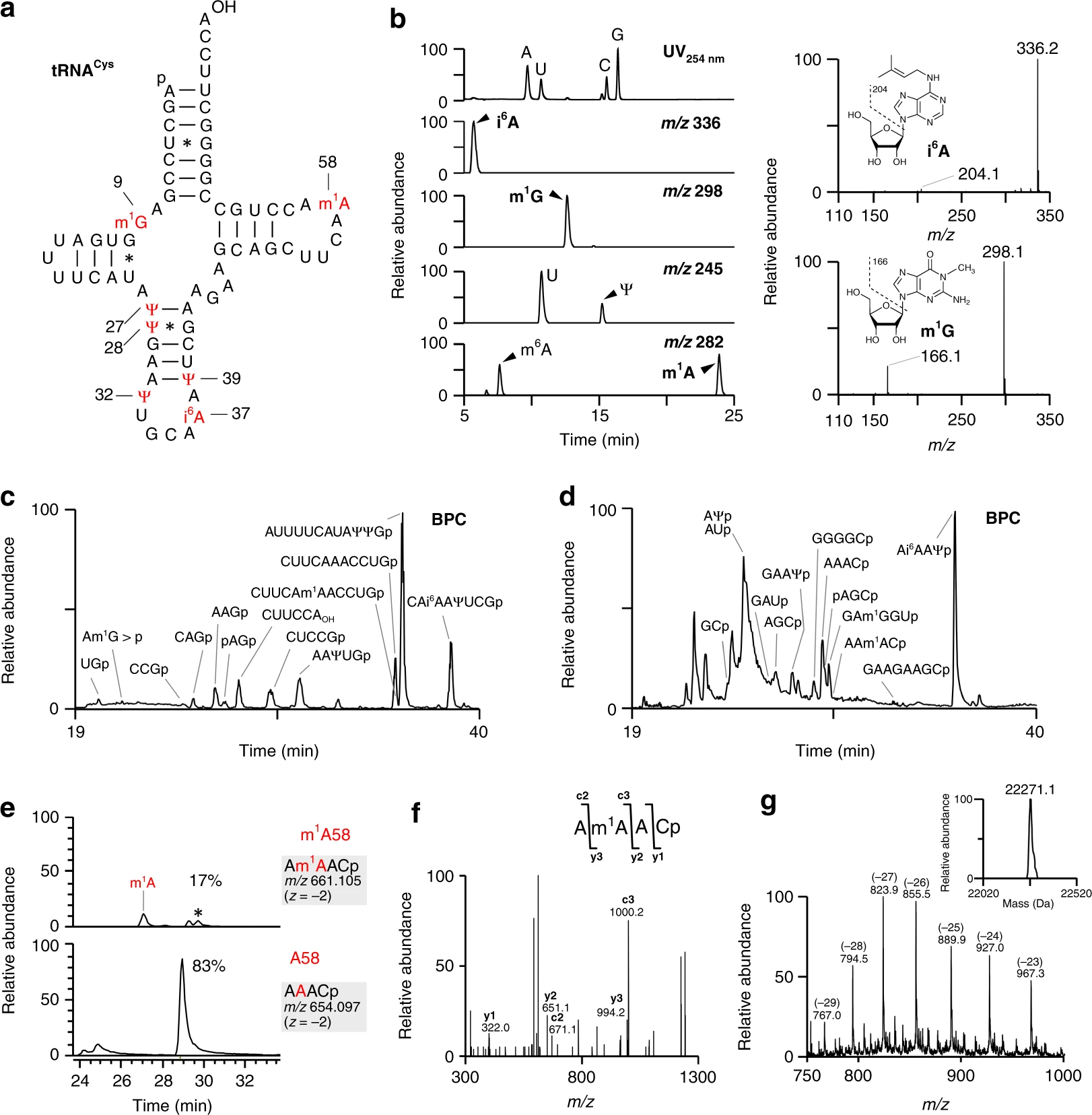
Suzuki Ta., Yashiro Y., Kikuchi I., Ishigami Y., Saito H., Matsuzawa I., Okada S., Mito M., Iwasaki S., Ma D., Zhao X., Asano K., Lin H., Kirino Y., Sakaguchi Y., and Suzuki Ts.*:
“Complete chemical structures of human mitochondrial tRNAs”
Nat Commun. 11(1):4269. (2020)
Press Release (RIKEN)
Press Release (The University of Tokyo)
Recommended in Faculty Opinions
“Complete chemical structures of human mitochondrial tRNAs”
Nat Commun. 11(1):4269. (2020)
Press Release (RIKEN)
Press Release (The University of Tokyo)
Recommended in Faculty Opinions

Nakazawa K. Shichino Y., Iwasaki S., and Shiina N.*:
“Implications of RNG140 (caprin2)-mediated translational regulation in eye lens differentiation”
J Biol Chem. 295(44):15029-15044. (2020)
Press Release (RIKEN)
Press Release (NIBB)
“Implications of RNG140 (caprin2)-mediated translational regulation in eye lens differentiation”
J Biol Chem. 295(44):15029-15044. (2020)
Press Release (RIKEN)
Press Release (NIBB)

Fujita T., Yokoyama T., Shirouzu M., Taguchi H., Ito T., and Iwasaki S.*:
“Anatomy of footprint extension in ribosome profiling reveals translational landscape mediated by S1 protein in bacteria"
Research Square. (2020)
“Anatomy of footprint extension in ribosome profiling reveals translational landscape mediated by S1 protein in bacteria"
Research Square. (2020)
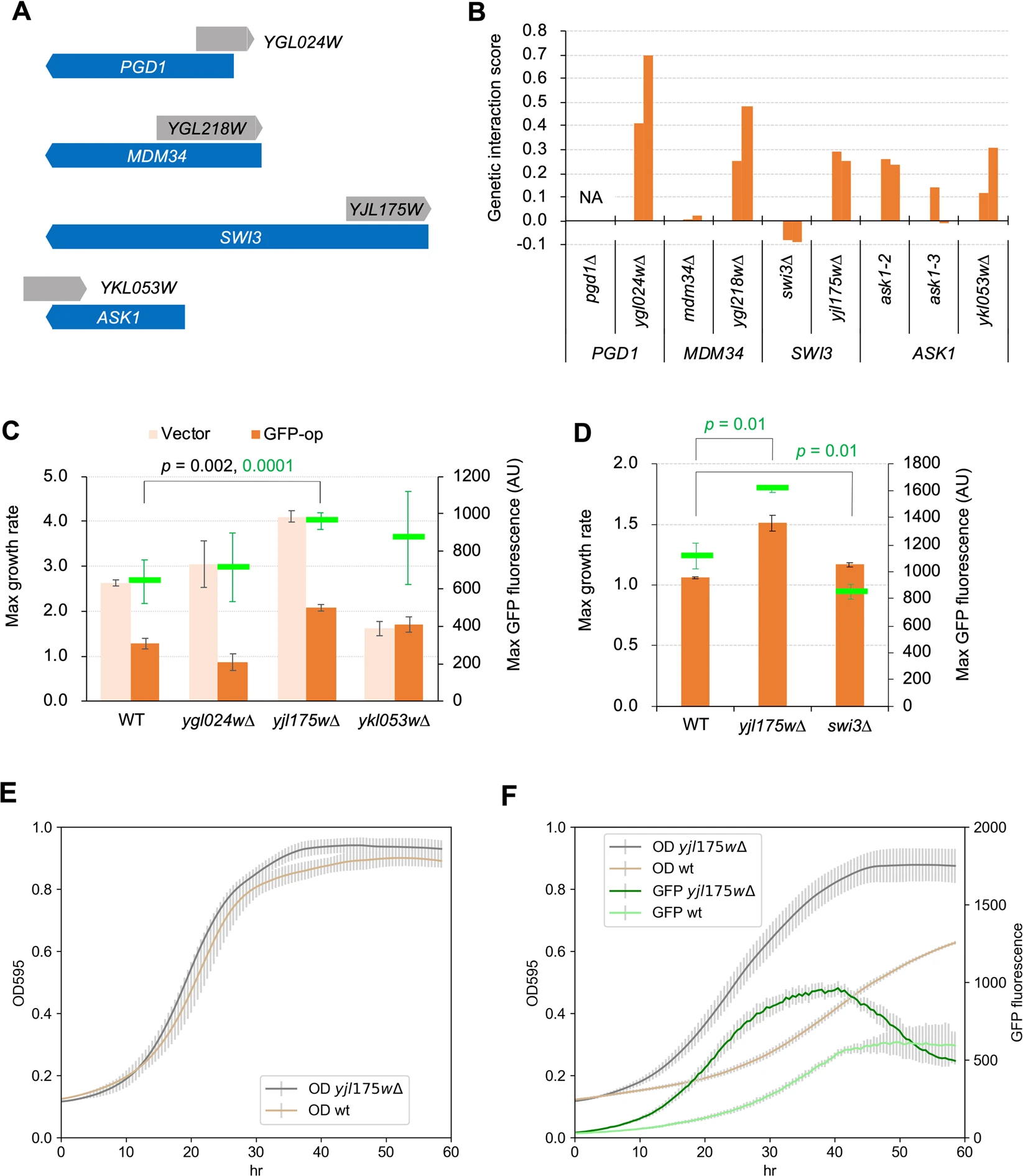
Saeki N., Eguchi Y., Kintaka R., Makanae K., Shichino Y., Iwasaki S., Kanno M., Kimura N., and Moriya H.*:
“N-terminal deletion of Swi3 created by the deletion of a dubious ORF YJL175W mitigates protein burden effect in S. cerevisiae”
Sci Rep. 10(1):9500. (2020)
“N-terminal deletion of Swi3 created by the deletion of a dubious ORF YJL175W mitigates protein burden effect in S. cerevisiae”
Sci Rep. 10(1):9500. (2020)

Han P., Shichino Y., Schneider-Poetsch T., Mito M., Hashimoto S., Udagawa T., Kohno K., Yoshida M., Mishima Y., Inada T., and Iwasaki S.*:
“Genome-wide survey of ribosome collision”
Cell Rep. 31(5):107610. (2020)
Addgene
BioRxiv. (2019)
Press Release
“Genome-wide survey of ribosome collision”
Cell Rep. 31(5):107610. (2020)
Addgene
BioRxiv. (2019)
Press Release
Tsuboyama K.*, Osaki T., Matsuura-Suzuki E., Kozuka-Hata H., Okada Y., Oyama M., Ikeuchi Y., Iwasaki S., and Tomari Y.*:
“A widespread family of heat-resistant obscure (Hero) proteins protect against protein instability and aggregation”
PLoS Biol. 18(3):e3000632. (2020)
BioRxiv. (2019)
Press Release (The University of Tokyo)
Media
“A widespread family of heat-resistant obscure (Hero) proteins protect against protein instability and aggregation”
PLoS Biol. 18(3):e3000632. (2020)
BioRxiv. (2019)
Press Release (The University of Tokyo)
Media

Yokoi S*, Naruse K, Kamei Y, Ansai S, Kinoshita M, Mito M, Iwasaki S., Inoue S, Okuyama T, Nakagawa S, Young LJ, and Takeuchi H.*:
“Sexually dimorphic role of oxytocin in medaka mate choice”
Proc Natl Acad Sci USA. 117(9):4802-4808. (2020)
“Sexually dimorphic role of oxytocin in medaka mate choice”
Proc Natl Acad Sci USA. 117(9):4802-4808. (2020)

Kurihara Y., Makita Y., Shimohira H., Fujita T., Iwasaki S., and Matsui M.*:
“Translational landscape of protein-coding and non-protein-coding RNAs upon light exposure in Arabidopsis”
Plant Cell Physiol. 61(3):536-545. (2020)
Press Release
“Translational landscape of protein-coding and non-protein-coding RNAs upon light exposure in Arabidopsis”
Plant Cell Physiol. 61(3):536-545. (2020)
Press Release

Hia F., Yang SF., Shichino Y., Yoshinaga M., Murakawa Y., Vandenbon A., Fukao A., Fujiwara T., Landthaler M., Natsume N., Adachi S., Iwasaki S., and Takeuchi O.*:
“Codon bias confers stability to human mRNAs”
EMBO Rep. 20(11):e48220. (2019)
BioRxiv. (2019)
“Codon bias confers stability to human mRNAs”
EMBO Rep. 20(11):e48220. (2019)
BioRxiv. (2019)

Padrón A., Iwasaki S., and Ingolia NT.*:
“Proximity RNA labeling by APEX-Seq reveals the organization of translation initiation complexes and repressive RNA granules”
Mol Cell. 75(4):875-887. (2019)
BioRxiv. (2018)
“Proximity RNA labeling by APEX-Seq reveals the organization of translation initiation complexes and repressive RNA granules”
Mol Cell. 75(4):875-887. (2019)
BioRxiv. (2018)

Hirayama H., Matsuda T., Tsuchiya Y., Oka R., Seino J., Huang C., Nakajima K., Noda Y., Shichino Y., Iwasaki S., and Suzuki T.*:
“Free glycans derived from O-mannosylated glycoproteins suggest the presence of an O-glycoprotein degradation pathway in yeast”
J Biol Chem. 294(44):15900-15911. (2019)
Selected as "Editors' Picks Highlights"
Press Release
“Free glycans derived from O-mannosylated glycoproteins suggest the presence of an O-glycoprotein degradation pathway in yeast”
J Biol Chem. 294(44):15900-15911. (2019)
Selected as "Editors' Picks Highlights"
Press Release

Fujita T., Kurihara Y.*, and Iwasaki S.*:
“The plant translatome surveyed by ribosome profiling”
Plant Cell Physiol. 60(9):1917-1926. (2019)
Selected as "Editor's choice"
“The plant translatome surveyed by ribosome profiling”
Plant Cell Physiol. 60(9):1917-1926. (2019)
Selected as "Editor's choice"
Mito M., Kadota M., Nakagawa S., and Iwasaki S.*:
“tChIP-Seq: cell-type-specific Eepigenome profiling”
J Vis Exp. 143:e58298. (2019)
“tChIP-Seq: cell-type-specific Eepigenome profiling”
J Vis Exp. 143:e58298. (2019)

Iwasaki S.*, Iwasaki W.#, Takahashi M.#, Sakamoto A., Watanabe C., Shichino Y., Floor SN., Fujiwara K., Mito M., Dodo K., Sodeoka M., Imataka H., Honma T., Fukuzawa K., Ito T.*, and Ingolia NT.*:
“The translation inhibitor rocaglamide targets a bimolecular cavity between eIF4A and polypurine RNA”
Mol Cell. 73(4):738-748.e9 (2019)
NBRP/RIKEN BRC
Press Release
Recommended in Faculty Opinions
“The translation inhibitor rocaglamide targets a bimolecular cavity between eIF4A and polypurine RNA”
Mol Cell. 73(4):738-748.e9 (2019)
NBRP/RIKEN BRC
Press Release
Recommended in Faculty Opinions

Akichika S.#, Hirano S.#, Shichino Y., Suzuki Ta., Nishimasu H., Ishitani R., Sugita A., Hirose Y., Iwasaki S., Nureki O.*, and Suzuki Ts.*:
“Cap-specific terminal N6-methylation of RNA by an RNA polymerase II–associated methyltransferase”
Science. 363(6423):eaav0080. (2019)
Press Release (RIKEN)
Press Release (The University of Tokyo)
Press Release (Nikkei)
Recommended in Faculty Opinions
“Cap-specific terminal N6-methylation of RNA by an RNA polymerase II–associated methyltransferase”
Science. 363(6423):eaav0080. (2019)
Press Release (RIKEN)
Press Release (The University of Tokyo)
Press Release (Nikkei)
Recommended in Faculty Opinions

Komatsu T., Yokoi S., Mito M., Fujii K., Kimura Y., Iwasaki S., and Nakagawa S.*:
“UPA-Seq: Prediction of functional lncRNAs using differential sensitivity to UV crosslinking”
RNA. 24(12):1785-1802. (2018)
BioRxiv (2018)
“UPA-Seq: Prediction of functional lncRNAs using differential sensitivity to UV crosslinking”
RNA. 24(12):1785-1802. (2018)
BioRxiv (2018)

Kurihara Y., Makita Y., Kawashima M., Fujita T., Iwasaki S., and Matsui M.*:
“Transcripts from downstream alternative transcription start sites evade uORF-mediated inhibition of gene expression in Arabidopsis”
Proc Natl Acad Sci USA. 115(30):7831-7836 (2018)
Highlighted by COMMENTARY
Press Release
Recommended in Faculty Opinions
“Transcripts from downstream alternative transcription start sites evade uORF-mediated inhibition of gene expression in Arabidopsis”
Proc Natl Acad Sci USA. 115(30):7831-7836 (2018)
Highlighted by COMMENTARY
Press Release
Recommended in Faculty Opinions
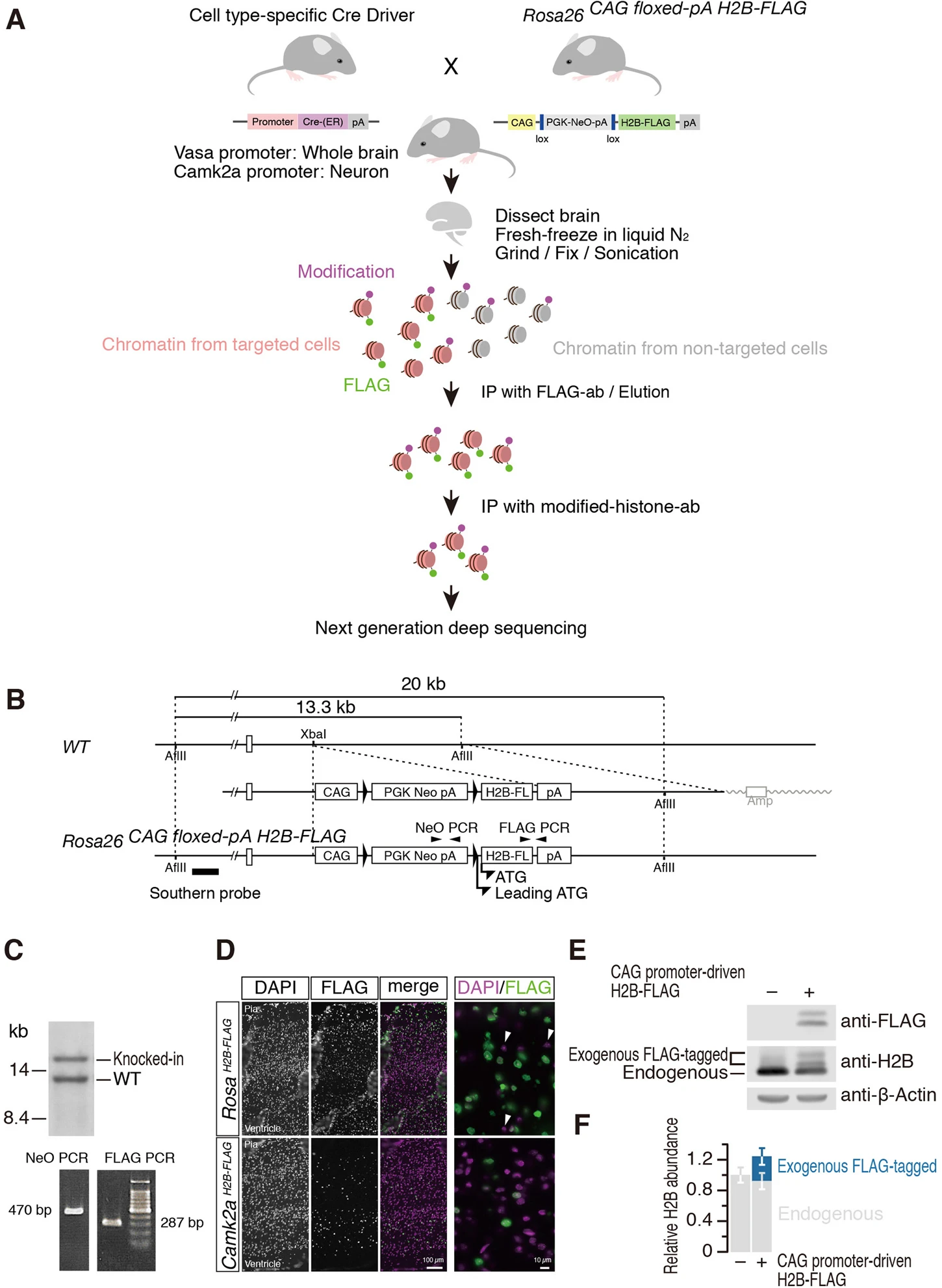
Mito M., Kadota M., Tanaka K., Furuta Y., Abe K., Iwasaki S.*, and Nakagawa S.*:
“Cell-type specific survey of epigenetic modifications by tandem chromatin immunoprecipitation sequencing”
Sci Rep. 8:1143. (2018)
BioRxiv. (2017)
Press Release
“Cell-type specific survey of epigenetic modifications by tandem chromatin immunoprecipitation sequencing”
Sci Rep. 8:1143. (2018)
BioRxiv. (2017)
Press Release

Iwasaki S. and Tomari Y.*:
“Reconstitution of RNA interference machinery”
Methods Mol Biol.1680:131-143. (2018)
“Reconstitution of RNA interference machinery”
Methods Mol Biol.1680:131-143. (2018)

Naruse K.#, Matsuura-Suzuki E.#, Watanabe M., Iwasaki S.*, and Tomari Y.*:
“In vitro reconstitution of chaperone-mediated human RISC assembly”
RNA. 24(1):6-11. (2018)
“In vitro reconstitution of chaperone-mediated human RISC assembly”
RNA. 24(1):6-11. (2018)
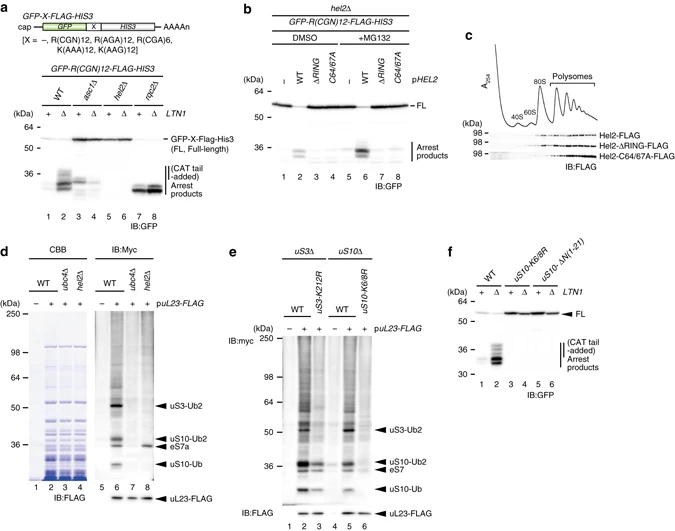
Matsuo Y.#, Ikeuchi K.#, Saeki, Y., Iwasaki S., Schmidt C., Udagawa T., Sato F., Tsuchiya H., Becker T., Tanaka K., Ingolia NT., Beckmann R., and Inada T.*:
“Ubiquitination of stalled ribosome triggers ribosome-associated quality control”
Nat Commun. 8(1):159. (2017)
“Ubiquitination of stalled ribosome triggers ribosome-associated quality control”
Nat Commun. 8(1):159. (2017)

Iwasaki S.* and Ingolia NT.*:
“The Growing toolbox for protein synthesis studies”
Trends Biochem Sci. 42(8):612-624. (2017)
“The Growing toolbox for protein synthesis studies”
Trends Biochem Sci. 42(8):612-624. (2017)

Ishikawa K., Makanae K., Iwasaki S., Ingolia NT., and Moriya H.*:
“Post-translational dosage compensation buffers genetic perturbations to stoichiometry of protein complexes”
PLoS Genet. 13(1):e1006554. (2017)
Press Release (Okayama University)
News (QLifePro)
“Post-translational dosage compensation buffers genetic perturbations to stoichiometry of protein complexes”
PLoS Genet. 13(1):e1006554. (2017)
Press Release (Okayama University)
News (QLifePro)

Iwasaki S. and Ingolia NT.*:
“PROTEIN TRANSLATION. Seeing translation”
Science. 352(6292):1391-2. (2016)
“PROTEIN TRANSLATION. Seeing translation”
Science. 352(6292):1391-2. (2016)

Iwasaki S., Floor SN., and Ingolia NT.*:
“Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor”
Nature. 534(7608):558-61. (2016)
NBRP/RIKEN BRC
“Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor”
Nature. 534(7608):558-61. (2016)
NBRP/RIKEN BRC

Werner A., Iwasaki S., Teerikorpi N., Fedrigo I., Ingolia NT., and Rape M.*:
“Cell-fate determination by ubiquitin-dependent regulation of translation”
Nature. 525(7570):523-7. (2015)
“Cell-fate determination by ubiquitin-dependent regulation of translation”
Nature. 525(7570):523-7. (2015)

Iwasaki S.#, Sasaki MH.#, Sakaguchi Y., Suzuki T., Tadakuma H.*, and Tomari Y.*:
“Defining fundamental steps in the assembly of Drosophila RNAi enzyme complex”
Nature. 521(7553):533-6. (2015)
Press Release
“Defining fundamental steps in the assembly of Drosophila RNAi enzyme complex”
Nature. 521(7553):533-6. (2015)
Press Release

Kwak PB.#, Iwasaki S.#, and Tomari Y.*:
“The microRNA pathway and cancer”
Cancer Sci. 101(11), 2309-15 (2010)
“The microRNA pathway and cancer”
Cancer Sci. 101(11), 2309-15 (2010)

Iwasaki S.#, Kobayashi M.#, Yoda M., Sakaguchi Y., Katsuma S., Suzuki T., and Tomari Y.*:
“Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes”
Mol Cell. 39(2):292-9. (2010)
Recommended in Faculty Opinions
“Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes”
Mol Cell. 39(2):292-9. (2010)
Recommended in Faculty Opinions

Yoda M., Kawamata T., Paroo Z., Ye X., Iwasaki S., Liu Q., and Tomari Y.*:
“ATP-dependent human RISC assembly pathways”
Nat Struct Mol Biol. 17(1):17-23. (2010)
“ATP-dependent human RISC assembly pathways”
Nat Struct Mol Biol. 17(1):17-23. (2010)

Iwasaki S., and Tomari Y.*:
“Argonaute-mediated translational repression (and activation)”
Fly (Austin). 3(3):204-6. (2009)
“Argonaute-mediated translational repression (and activation)”
Fly (Austin). 3(3):204-6. (2009)

Iwasaki S., Kawamata T., and Tomari Y.*:
“Drosophila Argonaute1 and Argonaute2 employ distinct mechanisms for translational repression”
Mol Cell. 34(1):58-67. (2009)
Journal Cover
Recommended in Faculty Opinions
“Drosophila Argonaute1 and Argonaute2 employ distinct mechanisms for translational repression”
Mol Cell. 34(1):58-67. (2009)
Journal Cover
Recommended in Faculty Opinions

Takeda A., Iwasaki S., Watanabe T., Utsumi M., and Watanabe Y.*:
“The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins”
Plant Cell Physiol. 49(4):493-500. (2008)
“The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins”
Plant Cell Physiol. 49(4):493-500. (2008)

Iwasaki S., Takeda A., Motose H., and Watanabe Y.*:
“Characterization of Arabidopsis decapping proteins AtDCP1 and AtDCP2, which are essential for post-embryonic development”
FEBS Lett. 581(13):2455-9. (2007)
“Characterization of Arabidopsis decapping proteins AtDCP1 and AtDCP2, which are essential for post-embryonic development”
FEBS Lett. 581(13):2455-9. (2007)

Japanese Reviews: 2025-current
前田佳穂, 岩崎信太郎:
"DAY2-2 遺伝情報の発現 ④~⑥":
7日間でプロになる 分子生物学これだけドリル (羊土社) (2025)
藤博貴, 山下映, 細金正樹, 岩崎信太郎:
“mRNA修飾によるトランスラトーム制御”
実験医学 43(11):1691-1697 (2025)
山下映, 岩崎信太郎:
“Ribo-Seqのデータ取得”
実験医学別冊 p49-65 (2025)
前田佳穂, 岩崎信太郎:
"DAY2-2 遺伝情報の発現 ④~⑥":
7日間でプロになる 分子生物学これだけドリル (羊土社) (2025)
藤博貴, 山下映, 細金正樹, 岩崎信太郎:
“mRNA修飾によるトランスラトーム制御”
実験医学 43(11):1691-1697 (2025)
山下映, 岩崎信太郎:
“Ribo-Seqのデータ取得”
実験医学別冊 p49-65 (2025)

Japanese Reviews: 2023-2024
脇川大誠, 岩崎信太郎:
“リボソームプロファイリングによるミトコンドリア翻訳の網羅的理解”
生化学 96(6):760-769. (2024)
岩崎信太郎:
“翻訳開始因子eIF4Aを標的とする翻訳阻害剤”
生化学 96(3):336-347. (2024)
戸室幸太郎, 藤博貴, 岩崎信太郎:
"リボソームプロファイリング法の拡張によるトランスレイトームの多面的理解"
実験医学増刊 41(15):2369-2375. (2023)
脇川大誠, 岩崎信太郎:
“リボソームプロファイリングによるミトコンドリア翻訳の網羅的理解”
生化学 96(6):760-769. (2024)
岩崎信太郎:
“翻訳開始因子eIF4Aを標的とする翻訳阻害剤”
生化学 96(3):336-347. (2024)
戸室幸太郎, 藤博貴, 岩崎信太郎:
"リボソームプロファイリング法の拡張によるトランスレイトームの多面的理解"
実験医学増刊 41(15):2369-2375. (2023)

Japanese Reviews: 2022
岩崎信太郎, 吉田稔:
"スプライシング阻害と翻訳制御"
生化学 94(6):819-828. (2022)
松浦絵里子, 岩崎信太郎:
"リボソームタンパク質の修飾と翻訳制御"
細胞 54(12):684-687. (2022)
市原知哉, 岩崎信太郎, 松本有樹修:
"未注釈ORFの同定とその生理機能"
実験医学増刊 40(12):2003-2010. (2022)
岩崎信太郎:
“ダイソームプロファイリング法によるリボソーム衝突の網羅探索”
生化学 94(1):87-91. (2022)
岩崎信太郎, 吉田稔:
"スプライシング阻害と翻訳制御"
生化学 94(6):819-828. (2022)
松浦絵里子, 岩崎信太郎:
"リボソームタンパク質の修飾と翻訳制御"
細胞 54(12):684-687. (2022)
市原知哉, 岩崎信太郎, 松本有樹修:
"未注釈ORFの同定とその生理機能"
実験医学増刊 40(12):2003-2010. (2022)
岩崎信太郎:
“ダイソームプロファイリング法によるリボソーム衝突の網羅探索”
生化学 94(1):87-91. (2022)

Japanese Reviews: 2017-2021
七野悠一, 岩崎信太郎:
“RNP顆粒研究を加速するトランスクリプトーム解析技術”
相分離生物学の全貌 (東京化学同人) 368-372. (2020)
木村悠介, 岩崎信太郎:
“リボソームプロファイリングによる網羅的翻訳解析の最前線”
実験医学 37(18):3055‐3057. (2019)
岩崎信太郎:
“eIF4A阻害の多様なメカニズム”
生化学 90(2):211‐215. (2018)
七野悠一, 岩崎信太郎:
“リボソームプロファイリングが切り拓く翻訳研究の未来”
実験医学 35(11):1868‐1873. (2017)
七野悠一, 岩崎信太郎:
“RNP顆粒研究を加速するトランスクリプトーム解析技術”
相分離生物学の全貌 (東京化学同人) 368-372. (2020)
木村悠介, 岩崎信太郎:
“リボソームプロファイリングによる網羅的翻訳解析の最前線”
実験医学 37(18):3055‐3057. (2019)
岩崎信太郎:
“eIF4A阻害の多様なメカニズム”
生化学 90(2):211‐215. (2018)
七野悠一, 岩崎信太郎:
“リボソームプロファイリングが切り拓く翻訳研究の未来”
実験医学 35(11):1868‐1873. (2017)

Japanese Reviews: 2010-2016
岩崎信太郎:
“小分子rocaglateは翻訳開始因子eIF4AをRNA配列に対し選択的な翻訳抑制因子に変える”
新着論文レビュー (2016)
岩崎信太郎:
“ncRNA判定法 (リボソームプロファイリング)”
ノンコーディングRNA RNA分子の全体像を俯瞰する (化学同人) 181-191. (2016)
岩崎信太郎, 佐々木浩, 泊幸秀:
“再構成系および1分子観察により同定されたショウジョウバエにおけるRISCの形成の過程”
新着論文レビュー (2015)
岩崎信太郎, 泊幸秀:
“small RNA二本鎖のATP依存的なRISC積み込み反応はHsc70/Hsp90シャペロンマシナリーによって媒介される”
新着論文レビュー (2010)
岩崎信太郎:
“小分子rocaglateは翻訳開始因子eIF4AをRNA配列に対し選択的な翻訳抑制因子に変える”
新着論文レビュー (2016)
岩崎信太郎:
“ncRNA判定法 (リボソームプロファイリング)”
ノンコーディングRNA RNA分子の全体像を俯瞰する (化学同人) 181-191. (2016)
岩崎信太郎, 佐々木浩, 泊幸秀:
“再構成系および1分子観察により同定されたショウジョウバエにおけるRISCの形成の過程”
新着論文レビュー (2015)
岩崎信太郎, 泊幸秀:
“small RNA二本鎖のATP依存的なRISC積み込み反応はHsc70/Hsp90シャペロンマシナリーによって媒介される”
新着論文レビュー (2010)

Essay:
“私のYMW”
日本RNA学会会報 (2025)
“My two cents”
日本生化学会 (2024)
“金言とセブンイレブンの冷やし中華”
日本RNA学会会報 (2020)
“受賞によせて”
日本RNA学会会報 (2019)
“私のYMW”
日本RNA学会会報 (2025)
“My two cents”
日本生化学会 (2024)
“金言とセブンイレブンの冷やし中華”
日本RNA学会会報 (2020)
“受賞によせて”
日本RNA学会会報 (2019)


